Evolutionary Origin of Insect–Wolbachia Nutritional Mutualism
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Metaproteogenomic Insights Beyond Bacterial Response to Naphthalene
ORIGINAL ARTICLE ISME Journal – Original article Metaproteogenomic insights beyond bacterial response to 5 naphthalene exposure and bio-stimulation María-Eugenia Guazzaroni, Florian-Alexander Herbst, Iván Lores, Javier Tamames, Ana Isabel Peláez, Nieves López-Cortés, María Alcaide, Mercedes V. del Pozo, José María Vieites, Martin von Bergen, José Luis R. Gallego, Rafael Bargiela, Arantxa López-López, Dietmar H. Pieper, Ramón Rosselló-Móra, Jesús Sánchez, Jana Seifert and Manuel Ferrer 10 Supporting Online Material includes Text (Supporting Materials and Methods) Tables S1 to S9 Figures S1 to S7 1 SUPPORTING TEXT Supporting Materials and Methods Soil characterisation Soil pH was measured in a suspension of soil and water (1:2.5) with a glass electrode, and 5 electrical conductivity was measured in the same extract (diluted 1:5). Primary soil characteristics were determined using standard techniques, such as dichromate oxidation (organic matter content), the Kjeldahl method (nitrogen content), the Olsen method (phosphorus content) and a Bernard calcimeter (carbonate content). The Bouyoucos Densimetry method was used to establish textural data. Exchangeable cations (Ca, Mg, K and 10 Na) extracted with 1 M NH 4Cl and exchangeable aluminium extracted with 1 M KCl were determined using atomic absorption/emission spectrophotometry with an AA200 PerkinElmer analyser. The effective cation exchange capacity (ECEC) was calculated as the sum of the values of the last two measurements (sum of the exchangeable cations and the exchangeable Al). Analyses were performed immediately after sampling. 15 Hydrocarbon analysis Extraction (5 g of sample N and Nbs) was performed with dichloromethane:acetone (1:1) using a Soxtherm extraction apparatus (Gerhardt GmbH & Co. -

Abstract Betaproteobacteria Alphaproteobacteria
Abstract N-210 Contact Information The majority of the soil’s biosphere containins biodiveristy that remains yet to be discovered. The occurrence of novel bacterial phyla in soil, as well as the phylogenetic diversity within bacterial phyla with few cultured representatives (e.g. Acidobacteria, Anne Spain Dr. Mostafa S.Elshahed Verrucomicrobia, and Gemmatimonadetes) have been previously well documented. However, few studies have focused on the Composition, Diversity, and Novelty within Soil Proteobacteria Department of Botany and Microbiology Department of Microbiology and Molecular Genetics novel phylogenetic diversity within phyla containing numerous cultured representatives. Here, we present a detailed University of Oklahoma Oklahoma State University phylogenetic analysis of the Proteobacteria-affiliated clones identified in a 13,001 nearly full-length 16S rRNA gene clones 770 Van Vleet Oval 307 LSE derived from Oklahoma tall grass prairie soil. Proteobacteria was the most abundant phylum in the community, and comprised Norman, OK 73019 Stillwater, OK 74078 25% of total clones. The most abundant and diverse class within the Proteobacteria was Alphaproteobacteria, which comprised 405 325 5255 405 744 6790 39% of Proteobacteria clones, followed by the Deltaproteobacteria, Betaproteobacteria, and Gammaproteobacteria, which made Anne M. Spain (1), Lee R. Krumholz (1), Mostafa S. Elshahed (2) up 37, 16, and 8% of Proteobacteria clones, respectively. Members of the Epsilonproteobacteria were not detected in the dataset. [email protected] [email protected] Detailed phylogenetic analysis indicated that 14% of the Proteobacteria clones belonged to 15 novel orders and 50% belonged (1) Dept. of Botany and Microbiology, University of Oklahoma, Norman, OK to orders with no described cultivated representatives or were unclassified. -

Anaplasmosis: an Emerging Tick-Borne Disease of Importance in Canada
IDCases 14 (2018) xxx–xxx Contents lists available at ScienceDirect IDCases journal homepage: www.elsevier.com/locate/idcr Case report Anaplasmosis: An emerging tick-borne disease of importance in Canada a, b,c d,e e,f Kelsey Uminski *, Kamran Kadkhoda , Brett L. Houston , Alison Lopez , g,h i c c Lauren J. MacKenzie , Robbin Lindsay , Andrew Walkty , John Embil , d,e Ryan Zarychanski a Rady Faculty of Health Sciences, Max Rady College of Medicine, Department of Internal Medicine, University of Manitoba, Winnipeg, MB, Canada b Cadham Provincial Laboratory, Government of Manitoba, Winnipeg, MB, Canada c Rady Faculty of Health Sciences, Max Rady College of Medicine, Department of Medical Microbiology and Infectious Diseases, University of Manitoba, Winnipeg, MB, Canada d Rady Faculty of Health Sciences, Max Rady College of Medicine, Department of Internal Medicine, Section of Medical Oncology and Hematology, University of Manitoba, Winnipeg, MB, Canada e CancerCare Manitoba, Department of Medical Oncology and Hematology, Winnipeg, MB, Canada f Rady Faculty of Health Sciences, Max Rady College of Medicine, Department of Pediatrics and Child Health, Section of Infectious Diseases, Winnipeg, MB, Canada g Rady Faculty of Health Sciences, Max Rady College of Medicine, Department of Internal Medicine, Section of Infectious Diseases, University of Manitoba, Winnipeg, MB, Canada h Rady Faculty of Health Sciences, Max Rady College of Medicine, Department of Community Health Sciences, University of Manitoba, Winnipeg, MB, Canada i Public Health Agency of Canada, National Microbiology Laboratory, Zoonotic Diseases and Special Pathogens, Winnipeg, MB, Canada A R T I C L E I N F O A B S T R A C T Article history: Human Granulocytic Anaplasmosis (HGA) is an infection caused by the intracellular bacterium Received 11 September 2018 Anaplasma phagocytophilum. -

Ehrlichiosis and Anaplasmosis Are Tick-Borne Diseases Caused by Obligate Anaplasmosis: Intracellular Bacteria in the Genera Ehrlichia and Anaplasma
Ehrlichiosis and Importance Ehrlichiosis and anaplasmosis are tick-borne diseases caused by obligate Anaplasmosis: intracellular bacteria in the genera Ehrlichia and Anaplasma. These organisms are widespread in nature; the reservoir hosts include numerous wild animals, as well as Zoonotic Species some domesticated species. For many years, Ehrlichia and Anaplasma species have been known to cause illness in pets and livestock. The consequences of exposure vary Canine Monocytic Ehrlichiosis, from asymptomatic infections to severe, potentially fatal illness. Some organisms Canine Hemorrhagic Fever, have also been recognized as human pathogens since the 1980s and 1990s. Tropical Canine Pancytopenia, Etiology Tracker Dog Disease, Ehrlichiosis and anaplasmosis are caused by members of the genera Ehrlichia Canine Tick Typhus, and Anaplasma, respectively. Both genera contain small, pleomorphic, Gram negative, Nairobi Bleeding Disorder, obligate intracellular organisms, and belong to the family Anaplasmataceae, order Canine Granulocytic Ehrlichiosis, Rickettsiales. They are classified as α-proteobacteria. A number of Ehrlichia and Canine Granulocytic Anaplasmosis, Anaplasma species affect animals. A limited number of these organisms have also Equine Granulocytic Ehrlichiosis, been identified in people. Equine Granulocytic Anaplasmosis, Recent changes in taxonomy can make the nomenclature of the Anaplasmataceae Tick-borne Fever, and their diseases somewhat confusing. At one time, ehrlichiosis was a group of Pasture Fever, diseases caused by organisms that mostly replicated in membrane-bound cytoplasmic Human Monocytic Ehrlichiosis, vacuoles of leukocytes, and belonged to the genus Ehrlichia, tribe Ehrlichieae and Human Granulocytic Anaplasmosis, family Rickettsiaceae. The names of the diseases were often based on the host Human Granulocytic Ehrlichiosis, species, together with type of leukocyte most often infected. -

Phylogenomics of the Reproductive Parasite Wolbachia Pipientis Wmel: a Streamlined Genome Overrun by Mobile Genetic Elements
PLoS BIOLOGY Phylogenomics of the Reproductive Parasite Wolbachia pipientis wMel: A Streamlined Genome Overrun by Mobile Genetic Elements Martin Wu1, Ling V. Sun2, Jessica Vamathevan1, Markus Riegler3, Robert Deboy1, Jeremy C. Brownlie3, Elizabeth A. McGraw3, William Martin4, Christian Esser4, Nahal Ahmadinejad4, Christian Wiegand4, Ramana Madupu1, Maureen J. Beanan1, Lauren M. Brinkac1, Sean C. Daugherty1, A. Scott Durkin1, James F. Kolonay1, William C. Nelson1, Yasmin Mohamoud1, Perris Lee1, Kristi Berry1, M. Brook Young1, Teresa Utterback1, Janice Weidman1, William C. Nierman1, Ian T. Paulsen1, Karen E. Nelson1, Herve´ Tettelin1, Scott L. O’Neill2,3, Jonathan A. Eisen1* 1 The Institute for Genomic Research, Rockville, Maryland, United States of America, 2 Department of Epidemiology and Public Health, Yale University School of Medicine, New Haven, Connecticut, United States of America, 3 Department of Zoology and Entomology, School of Life Sciences, The University of Queensland, St Lucia, Queensland, Australia, 4 Institut fu¨r Botanik III, Heinrich-Heine Universita¨t, Du¨sseldorf, Germany The complete sequence of the 1,267,782 bp genome of Wolbachia pipientis wMel, an obligate intracellular bacteria of Drosophila melanogaster, has been determined. Wolbachia, which are found in a variety of invertebrate species, are of great interest due to their diverse interactions with different hosts, which range from many forms of reproductive parasitism to mutualistic symbioses. Analysis of the wMel genome, in particular phylogenomic comparisons with other intracellular bacteria, has revealed many insights into the biology and evolution of wMel and Wolbachia in general. For example, the wMel genome is unique among sequenced obligate intracellular species in both being highly streamlined and containing very high levels of repetitive DNA and mobile DNA elements. -

Acidification Increases Abundances of Vibrionales And
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Sapientia Acidification increases abundances of Vibrionales and Planctomycetia associated to a seaweed-grazer system: potential consequences for disease and prey digestion efficiency Tania Aires1,*, Alexandra Serebryakova1,2,*, Frédérique Viard2,3, Ester A. Serrão1 and Aschwin H. Engelen1 1 Center for Marine Sciences (CCMAR), CIMAR, University of Algarve, Campus de Gambelas, Faro, Portugal 2 Sorbonne Université, CNRS, Lab Adaptation and Diversity in Marine Environments (UMR 7144 CNRS SU), Station Biologique de Roscoff, Roscoff, France 3 CNRS, UMR 7144, Divco Team, Station Biologique de Roscoff, Roscoff, France * These authors contributed equally to this work. ABSTRACT Ocean acidification significantly affects marine organisms in several ways, with complex interactions. Seaweeds might benefit from rising CO2 through increased photosynthesis and carbon acquisition, with subsequent higher growth rates. However, changes in seaweed chemistry due to increased CO2 may change the nutritional quality of tissue for grazers. In addition, organisms live in close association with a diverse microbiota, which can also be influenced by environmental changes, with feedback effects. As gut microbiomes are often linked to diet, changes in seaweed characteristics and associated microbiome can affect the gut microbiome of the grazer, with possible fitness consequences. In this study, we experimentally investigated the effects of acidification on the microbiome of the invasive brown seaweed Sargassum muticum and a native isopod consumer Synisoma nadejda. Both were exposed to ambient CO2 conditions Submitted 13 September 2017 (380 ppm, pH 8.16) and an acidification treatment (1,000 ppm, pH 7.86) for three Accepted 26 January 2018 weeks. -

Legionella Shows a Diverse Secondary Metabolism Dependent on a Broad Spectrum Sfp-Type Phosphopantetheinyl Transferase
Legionella shows a diverse secondary metabolism dependent on a broad spectrum Sfp-type phosphopantetheinyl transferase Nicholas J. Tobias1, Tilman Ahrendt1, Ursula Schell2, Melissa Miltenberger1, Hubert Hilbi2,3 and Helge B. Bode1,4 1 Fachbereich Biowissenschaften, Merck Stiftungsprofessur fu¨r Molekulare Biotechnologie, Goethe Universita¨t, Frankfurt am Main, Germany 2 Max von Pettenkofer Institute, Ludwig-Maximilians-Universita¨tMu¨nchen, Munich, Germany 3 Institute of Medical Microbiology, University of Zu¨rich, Zu¨rich, Switzerland 4 Buchmann Institute for Molecular Life Sciences, Goethe Universita¨t, Frankfurt am Main, Germany ABSTRACT Several members of the genus Legionella cause Legionnaires’ disease, a potentially debilitating form of pneumonia. Studies frequently focus on the abundant number of virulence factors present in this genus. However, what is often overlooked is the role of secondary metabolites from Legionella. Following whole genome sequencing, we assembled and annotated the Legionella parisiensis DSM 19216 genome. Together with 14 other members of the Legionella, we performed comparative genomics and analysed the secondary metabolite potential of each strain. We found that Legionella contains a huge variety of biosynthetic gene clusters (BGCs) that are potentially making a significant number of novel natural products with undefined function. Surprisingly, only a single Sfp-like phosphopantetheinyl transferase is found in all Legionella strains analyzed that might be responsible for the activation of all carrier proteins in primary (fatty acid biosynthesis) and secondary metabolism (polyketide and non-ribosomal peptide synthesis). Using conserved active site motifs, we predict Submitted 29 June 2016 some novel compounds that are probably involved in cell-cell communication, Accepted 25 October 2016 Published 24 November 2016 differing to known communication systems. -

Ultrastructure and Localization of Neorickettsia in Adult Digenean
Washington University School of Medicine Digital Commons@Becker Open Access Publications 2017 Ultrastructure and localization of Neorickettsia in adult digenean trematodes provides novel insights into helminth-endobacteria interaction Kerstin Fischer Washington University School of Medicine in St. Louis Vasyl V. Tkach University of North Dakota Kurt C. Curtis Washington University School of Medicine in St. Louis Peter U. Fischer Washington University School of Medicine in St. Louis Follow this and additional works at: https://digitalcommons.wustl.edu/open_access_pubs Recommended Citation Fischer, Kerstin; Tkach, Vasyl V.; Curtis, Kurt C.; and Fischer, Peter U., ,"Ultrastructure and localization of Neorickettsia in adult digenean trematodes provides novel insights into helminth-endobacteria interaction." Parasites & Vectors.10,. 177. (2017). https://digitalcommons.wustl.edu/open_access_pubs/5789 This Open Access Publication is brought to you for free and open access by Digital Commons@Becker. It has been accepted for inclusion in Open Access Publications by an authorized administrator of Digital Commons@Becker. For more information, please contact [email protected]. Fischer et al. Parasites & Vectors (2017) 10:177 DOI 10.1186/s13071-017-2123-7 RESEARCH Open Access Ultrastructure and localization of Neorickettsia in adult digenean trematodes provides novel insights into helminth- endobacteria interaction Kerstin Fischer1, Vasyl V. Tkach2, Kurt C. Curtis1 and Peter U. Fischer1* Abstract Background: Neorickettsia are a group of intracellular α proteobacteria transmitted by digeneans (Platyhelminthes, Trematoda). These endobacteria can also infect vertebrate hosts of the helminths and cause serious diseases in animals and humans. Neorickettsia have been isolated from infected animals and maintained in cell cultures, and their morphology in mammalian cells has been described. -

The Risk to Human Health from Free-Living Amoebae Interaction with Legionella in Drinking and Recycled Water Systems
THE RISK TO HUMAN HEALTH FROM FREE-LIVING AMOEBAE INTERACTION WITH LEGIONELLA IN DRINKING AND RECYCLED WATER SYSTEMS Dissertation submitted by JACQUELINE MARIE THOMAS BACHELOR OF SCIENCE (HONOURS) AND BACHELOR OF ARTS, UNSW In partial fulfillment of the requirements for the award of DOCTOR OF PHILOSOPHY in ENVIRONMENTAL ENGINEERING SCHOOL OF CIVIL AND ENVIRONMENTAL ENGINEERING FACULTY OF ENGINEERING MAY 2012 SUPERVISORS Professor Nicholas Ashbolt Office of Research and Development United States Environmental Protection Agency Cincinnati, Ohio USA and School of Civil and Environmental Engineering Faculty of Engineering The University of New South Wales Sydney, Australia Professor Richard Stuetz School of Civil and Environmental Engineering Faculty of Engineering The University of New South Wales Sydney, Australia Doctor Torsten Thomas School of Biotechnology and Biomolecular Sciences Faculty of Science The University of New South Wales Sydney, Australia ORIGINALITY STATEMENT '1 hereby declare that this submission is my own work and to the best of my knowledge it contains no materials previously published or written by another person, or substantial proportions of material which have been accepted for the award of any other degree or diploma at UNSW or any other educational institution, except where due acknowledgement is made in the thesis. Any contribution made to the research by others, with whom 1 have worked at UNSW or elsewhere, is explicitly acknowledged in the thesis. I also declare that the intellectual content of this thesis is the product of my own work, except to the extent that assistance from others in the project's design and conception or in style, presentation and linguistic expression is acknowledged.' Signed ~ ............................ -

18167638.Pdf
REVIEW ARTICLE Amoebal pathogens as emerging causal agents of pneumonia Fred´ eric´ Lamoth1 & Gilbert Greub1,2 1Infectious Diseases Service, University of Lausanne, Lausanne, Switzerland; and 2Center for Research on Intracellular Bacteria, Institute of Microbiology, University Hospital Center and University of Lausanne, Lausanne, Switzerland Correspondence: Gilbert Greub, Center for Abstract Research on Intracellular Bacteria, Institute of Microbiology, University Hospital Center and Despite using modern microbiological diagnostic approaches, the aetiological University of Lausanne, Rue du Bugnon 46, agents of pneumonia remain unidentified in about 50% of cases. Some bacteria 1011 Lausanne, Switzerland. Tel.: 141 21 that grow poorly or not at all in axenic media used in routine clinical bacteriology 31449 79; fax: 141 21 31440 60; e-mail: laboratory but which can develop inside amoebae may be the agents of these lower [email protected] respiratory tract infections (RTIs) of unexplained aetiology. Such amoebae- resisting bacteria, which coevolved with amoebae to resist their microbicidal Received 24 September 2009; revised 30 machinery, may have developed virulence traits that help them survive within November 2009; accepted 2 December 2009. human macrophages, i.e. the first line of innate immune defence in the lung. We Final version published online 22 January 2010. review here the current evidence for the emerging pathogenic role of various DOI:10.1111/j.1574-6976.2009.00207.x amoebae-resisting microorganisms as agents of RTIs in humans. Specifically, we discuss the emerging pathogenic roles of Legionella-like amoebal pathogens, novel Editor: Colin Berry Chlamydiae (Parachlamydia acanthamoebae, Simkania negevensis), waterborne mycobacteria and Bradyrhizobiaceae (Bosea and Afipia spp.). Keywords free-living amoebae; amoebae-resisting bacteria; Legionella; Chlamydia-like bacteria; mycobacteria; pneumonia. -
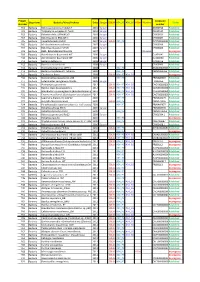
Project Number Organisms Bacteria/Virus/Archaea Date
Project_ Accession Organisms Bacteria/Virus/Archaea Date Sanger SOLiD 454_PE 454_SG PGM Illumina Status Number number P01 Bacteria Rickettsia conorii str.Malish 7 2001 Sanger AE006914 Published P02 Bacteria Tropheryma whipplei str.Twist 2003 Sanger AE014184 Published P03 Bacteria Rickettsia felis URRWXCal2 2005 Sanger CP000053 Published P04 Bacteria Rickettsia bellii RML369-C 2006 Sanger CP000087 Published P05 Bacteria Coxiella burnetii CB109 2007 Sanger SOLiD 454_PE AKYP00000000 Published P06 Bacteria Minibacterium massiliensis 2007 Sanger CP000269 Published P07 Bacteria Rickettsia massiliae MTU5 2007 Sanger CP000683 Published P08 Bacteria BaBL=Bête à Bernard Lascola 2007 Illumina In progress P09 Bacteria Acinetobacter baumannii AYE 2006 Sanger CU459141 Published P10 Bacteria Acinetobacter baumannii SDF 2006 Sanger CU468230 Published P11 Bacteria Borrelia duttonii Ly 2008 Sanger CP000976 Published P12 Bacteria Borrelia recurrentis A1 2008 Sanger CP000993 Published P13 Bacteria Francisella tularensis URFT1 2008 454_PE ABAZ00000000Published P14 Bacteria Borrelia crocidurae str. Achema 2009 454_PE PRJNA162335 Published P15 Bacteria Citrobacter koseri 2009 SOLiD 454_PE 454_SG In progress P16 Bacteria Diplorickettsia massiliensis 20B 2009 454_PE PRJNA86907 Published P17 Bacteria Enterobacter aerogenes EA1509E 2009 Sanger FO203355 Published P18 Bacteria Actinomyces grossensis 2012 SOLiD 454_PE 454_SG CAGY00000000Published P19 Bacteria Bacillus massiliosenegalensis 2012 SOLiD 454_PE 454_SG CAHJ00000000 Published P20 Bacteria Brevibacterium senegalensis -

ESCMID Online Lecture Library © by Author
The order Rickettsiales Pathogenesis of “ehrlichia” (Anaplasmataceae) infections of humans •Family Rickettsiaceae • Genera Rickettsia, Orientia •Family Anaplasmatacea • Ehrlichia, Anaplasma, others J. Stephen Dumler, M.D. •obligate intracellular bacteria • -proteobacteria phylogeny by small Departments of Pathology and Microbiology & Immunology subunit RNA genes (rrs) University of Maryland School of Medicine • contain DNA, RNA, ribosomes And Departments of Pathology and Molecular Microbiology & Immunology • divide by binary fission The Johns Hopkins Medical Institutions • Gram-negative cell wall Baltimore, MD USA •life cycle within arthropod host [email protected] •Bartonella (and former Rochalimaea) and Coxiella not in Rickettsiales Phylogeny of Rickettsiales (rrs) Ehrlichia and Anaplasma species infections α-proteobacteria E chaffeensis E ewingii pathogenesis A phagocytophilum E muris R prowazekii R australis • attachment R typhi R akari R felis R honei • endosomal entry R rickettsii Neoehrlichia mikurensis R conorii R parkeri W pipientis R africae R sibirica • avoidance of immediate intracellular killing (signaling) – phagolysosome fusion inhibition - doxycycline sensitive O tsutsugamushi Anaplasmataceae – subversion of autophagy Rickettsiaceae – detoxification of oxidative killing mechanisms • regulation and perturbation of host cell functions N sennetsu (gene expression) – downregulation of innate or early immune responses B quintana – cell cycle perturbations / apoptosis B henselae E coli – inhibition of intracellular trafficking