An Immuno-Assay to Quantify Influenza Virus Hemagglutinin with Correctly Folded Stalk Domains in Vaccine Preparations
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Laboratory Techniques Used for Immunological Laboratory Methods
Laboratory techniques used for Immunological laboratory methods Dr. Tatiana Jones, MD, PhD NCC How to Make Serial Dilutions? Interpretation can be made differently depending on the nature of test. For example, if we need to figure out in what sample the concentration of the antibody or antigen is higher, we will go by TITER, which is the lowest serial dilution (let’s say that it is 1:32 in the picture on the left) that gives us positive result. This mean that even diluted 32 times sample is still capable of reacting. The other scenario when we are interpreting quantitative assays, such as ELISA. In this case we need to match results of our samples to known concentrations of STANDARD and MULTIPLY be our dilution factor. What is Antibody Titer? An antibody titer is a measurement of how much antibody an organism has produced that recognizes a particular antigen. Titer is expressed as the inverse of the greatest dilution that still gives a positive result. ELISA is a common means of determining antibody titers. How to Determine Antibody Titer? Where we can use Indirect Coombs test detects the presence of anti-Rh antibodies in blood serum. A patient might be reported to have an "indirect Antibody Titer? Coombs titer" of 16. This means that the patient's serum gives a positive indirect Coombs test at any dilution down to 1/16 (1 part serum to 15 parts diluent). At greater dilutions the indirect Coombs test is negative. If a few weeks later the same patient had an indirect Coombs titer of 32 (1/32 dilution which is 1 part serum to 31 parts diluent), this would mean that more anti-Rh antibody was made, since it took a greater dilution to eradicate the positive test. -

Radial Immunodiffusion Assay Protocol
Radial Immunodiffusion Aim: To study the immunodiffusion technique by Single Radial Immunodiffusion. Introduction: Single Radial Immunodiffusion, also known as Mancini technique, is a quantitative immunodiffusion technique used to detect the concentration of antigen by measuring the diameter of the precipitin ring formed by the interaction of the antigen and the antibody at optimal concentration. In this method the antibody is incorporated into the agarose gel whereas the antigen diffuses into it in a radial pattern. Thus, the antibody is uniformly distributed throughout the gel. Principle: Single Radial Immunodiffusion is used extensively for the quantitative estimation of antigen. Here the antigen-antibody reaction is made more sensitive by the addition of antiserum into the agarose gel and loading the antigen sample in the well. As the antigen diffuses into the agarose radially in all directions, it’s concentration continuously falls until the equivalence point is reached at which the antigen concentration is in equal proportion to that of the antibody present in the agarose gel. At this point ring of precipitation (‘precipitin ring’) is formed around the well. The diameter of the precipitin ring is proportional to the concentration of antigen. With increasing concentration of antigen, precipitin rings with larger diameter are formed. The size of the precipitin rings depends on: Antigen concentration in the sample well Antibody concentration in the agarose gel Size of the sample well Volume of the sample Thus, by having various concentrations of a standard antigen, standard curve can be obtained from which one can determine the amount of an antigen in an unknown sample. Thus, this is a quantitative test. -

Importance of Ag-Ab Reactions
Ag-Ab reactions Tests for Ag-Ab reactions EISA SALEHI PhD. Immunology Dept. TUMS Importance of Ag-Ab Reactions • Understand the mechanisms of defense • Abs as tools in: – Treatment – Diagnosis • As biomarkers • As tools to measure analytes Nature of Ag/Ab Reactions http://www.med.sc.edu:85/chime2/lyso-abfr.htm • Lock and Key Concept • Non-covalent Bonds – Hydrogen bonds – Electrostatic bonds – Van der Waal forces – Hydrophobic bonds • Multiple Bonds • Reversible Source: Li, Y., Li, H., Smith-Gill, S. J., Mariuzza, R. A., Biochemistry 39, 6296, 2000 Affinity • Strength of the reaction between a single antigenic determinant and a single Ab combining site High Affinity Low Affinity Ab Ab Ag Ag Affinity = ( attractive and repulsive forces Calculation of Affinity Ag + Ab ↔ Ag-Ab Applying the Law of Mass Action: [[gAg-Ab] Keq = [Ag] x [Ab] Avidity • The overall strength of binding between an Ag with many determinants and multivalent Abs 4 6 10 Keq = 10 10 10 Affinity Avidity Avidity SifiitSpecificity • The ability of an individual antibody combining site to react with only one antigenic determinant. • The ability of a population of antibody molecules to react with only one antigen. Cross Reactivity • The ability of an individual Ab combining site to react with more than one antigenic determinant. • The ability of a population of Ab molecules to react with more than one Ag Cross reactions Anti-A Anti-A Anti-A Ab Ab Ab Ag A Ag B Ag C Shared epitope Similar epitope Factors Affecting Measurement of A/AbRAg/Ab Reac tions • Affinity • Avidity Ab excess Ag excess • AAbiAg:Ab ratio •Phyygsical form of Ag Equivalence – Lattice formation Do you need to know what happens in Lab. -

Diagnostic Potential of the Haemagglutination
Bulgarian Journal of Veterinary Medicine (2007), 10 , N o 3, 169 −178 DIAGNOSTIC POTENTIAL OF THE HAEMAGGLUTINATION INHIBITION TEST, THE IMMUNODIFFUSION TEST AND ELISA FOR DETECTION OF ANTIBODIES IN CHICKENS, INTRAVENOUSLY INFECTED WITH A/DUCK/BULGARIA/05 H6N2 AVIAN INFLUENZA VIRUS ISOLATE I. S. ZARKOV Faculty of Veterinary Medicine, Trakia University, Stara Zagora, Bulgaria Summary Zarkov, I., 2007. Diagnostic potential of the haemagglutination inhibition test, the immuno- diffusion test and ELISA for detection of antibodies in chickens, intravenously infected with A/duck/Bulgaria/05 H6N2 avian influenza virus isolate. Bulg. J. Vet. Med. , 10 , No 3, 169 −178. After experimental infection of chickens with an avian influenza viral isolate A/duck/Bulgaria/05 H6N2, the potential of the haemagglutination inhibition test (HIT), the immunodiffusion test (IDT) and enzyme-linked immunosorbent assay (ELISA) for antibody detection was evaluated. The results evidenced that in birds, haemagglutinins, precipitins and IgG antibodies, detectable with ELISA, were formed. The percentage of chickens with subtype-specific antibodies was the highest by the 21 st day of infection (100 %), followed by the 14 th (66.7 %) and the 28 th (55.6 %) days, and was the lowest by the 7 th day (44.4 %). Serum titres ranged between 1:4 and 1:256 with predomination of 1:8 and 1:16 titres (29.2 % each). The mean arithmetic titre for the experiment was 1:38.2. The highest percentage of chickens with precipitins was observed by the 14th day (55.6 %) followed by the 7 th , 21 st and the 28 th days with 33.3 % each. -

Serological Methods in the Identification and Characterization of Viruses
CHAPTER 4 Serological Methods in the Identification and Characterization of Viruses M. H. V. Van Regenmortel Laboratoire de Virologie Institut de Biologie Mo!eculaire et Cellulaire 67000 Strasbourg, France 1. INTRODUCTION The purpose of this chapter is to present an integrated view of the various serological techniques that have been used in virology. The accent will be placed on the principles that govern each type of test and on the general applicability of the different serological techniques in all fields of virus research. In recent years, advances in serological tech niques have sometimes been applied in only one area of virology, although they could have been equally useful to workers studying other groups of viruses. No doubt this stems from the host-oriented approach that has guided the compartmentation of virology into separate fields of specialization. When it comes to serological properties, however, the similarities between animal, insect, bacterial, and plant viruses are paramount. The same immunochemical principles govern the in vitro serological reactions of all viral antigens, and much of general interest can be learned from the findings obtained with each particular group of viruses. An attempt will be made here to emphasize the general validity of specific experimental procedures. A number of recent reviews restricted to the serology of particular groups of viruses are available 183 H. Fraenkel-Conrat et al. (eds.), Comprehensive Virology © Plenum Press, New York 1981 184 Chapter 4 (Cowan, 1973; Schmidt and Lennette, 1973; Ball, 1974; Kurstak and Morisset, 1974; Burns and Allison, 1975; Mazzone and Tignor, 1976; Mayr et al., 1977; Tyrrell, 1978; Van Regenmortel, 1978; Cooper, 1979). -

MCB 407 – Immunology and Immunochemistry COURSE PARTICULARS COURSE INSTRUCTORS COURSE DESCRIPTION
D DEPARTMENT OF MICROBIOLOGY MCB 407 – Immunology and Immunochemistry COURSE PARTICULARS Course Code: MCB 407 Course Title: Immunology and Immunochemistry No. of Units: 4 Course Duration: Three hours of theory and three hours of practical per week for 15 weeks. Status: Compulsory Course Email Address: [email protected] Course Webpage: http://www.fwt.futa.edu.ng/courseschedule.php?coursecode=MCB%407 Prerequisite: BIO 201, BCH 201 COURSE INSTRUCTORS Professor (Mrs). T. T. Adebolu Microbiology Office Annex, Room 14 Dept. of Microbiology, Federal University of Technology, Akure, Nigeria. Phone: +2348053617571 Email: [email protected] and Dr. M. K. Oladunmoye Postgraduate Research Laboratory Phase 1, Dept. of Microbiology, Federal University of Technology, Akure, Nigeria. Phone: +2348035057977 Email: [email protected] COURSE DESCRIPTION Basic concept of Immunology. Antigens and antigenic determinants. Antibodies. Structures and classification of immunoglobulins/antibodies. Antigen and antibody reactions. Innate and Acquired Immunity. Immune response. Hypersensitivity reactions. Autoimmune diseases. Immunodeficiency diseases. Introduction to transplantation immunology. The practicals will include laboratory exercise in modern techniques in immunology and immunochemistry. 1 COURSE OBJECTIVES The objectives of this course are to: give the students an insight to the basic concept of immunology; expose the students to the major determinants that confer immunity in a host to infections; and acquire practical skills for immunodiagnosis -

IMMUNOCHEMICAL TECHNIQUES Antigens Antibodies
Imunochemical Techniques IMMUNOCHEMICAL TECHNIQUES (by Lenka Fialová, translated by Jan Pláteník a Martin Vejražka) Antigens Antigens are macromolecules of natural or synthetic origin; chemically they consist of various polymers – proteins, polypeptides, polysaccharides or nucleoproteins. Antigens display two essential properties: first, they are able to evoke a specific immune response , either cellular or humoral type; and, second, they specifically interact with products of this immune response , i.e. antibodies or immunocompetent cells. A complete antigen – immunogen – consists of a macromolecule that bears antigenic determinants (epitopes) on its surface (Fig. 1). The antigenic determinant (epitope) is a certain group of atoms on the antigen surface that actually interacts with the binding site on the antibody or lymphocyte receptor for the antigen. Number of epitopes on the antigen surface determines its valency. Low-molecular-weight compound that cannot as such elicit production of antibodies, but is able to react specifically with the products of immune response, is called hapten (incomplete antigen) . antigen epitopes Fig. 1. Antigen and epitopes Antibodies Antibodies are produced by plasma cells that result from differentiation of B lymphocytes following stimulation with antigen. Antibodies are heterogeneous group of animal glycoproteins with electrophoretic mobility β - γ, and are also called immunoglobulins (Ig) . Every immunoglobulin molecule contains at least two light (L) and two heavy (H) chains connected with disulphidic bridges (Fig. 2). One antibody molecule contains only one type of light as well as heavy chain. There are two types of light chains - κ and λ - that determine type of immunoglobulin molecule; while heavy chains exist in 5 isotypes - γ, µ, α, δ, ε; and determine class of immunoglobulins - IgG, IgM, IgA, IgD and IgE . -

Virology Techniques
Chapter 5 - Lesson 4 Virology Techniques Introduction Virology is a field within microbiology that encom- passes the study of viruses and the diseases they cause. In the laboratory, viruses have served as useful tools to better understand cellular mechanisms. The purpose of this lesson is to provide a general overview of laboratory techniques used in the identification and study of viruses. A Brief History In the late 19th century the independent work of Dimitri Ivanofsky and Martinus Beijerinck marked the begin- This electron micrograph depicts an influenza virus ning of the field of virology. They showed that the agent particle or virion. CDC. responsible for causing a serious disease in tobacco plants, tobacco mosaic virus, was able to pass through filters known to retain bacteria and the filtrate was able to cause disease in new plants. In 1898, Friedrich Loef- fler and Paul Frosch applied the filtration criteria to a disease in cattle known as foot and mouth disease. The filtration criteria remained the standard method used to classify an agent as a virus for nearly 40 years until chemical and physical studies revealed the structural basis of viruses. These attributes have become the ba- sis of many techniques used in the field today. Background All organisms are affected by viruses because viruses are capable of infecting and causing disease in all liv- ing species. Viruses affect plants, humans, and ani- mals as well as bacteria. A virus that infects bacteria is known as a bacteriophage and is considered the Bacteriophage. CDC. Chapter 5 - Human Health: Real Life Example (Influenza) 1 most abundant biological entity on the planet. -

Interpretation of Serologic Tests
COCCIDIOIDOMYCOSIS SEROLOGY LABORATORY School of Medicine, University of California, Davis, California, 95616-8645 George Thompson M.D., Director NPI: 1831206812 VETERINARY CLIENTS Application and Interpretation of Serologic Tests A. Our usual serologic tests for coccidioidomycosis are the following: 1. Qualitative: Immunodiffusion (ID) to determine presence of coccidioidal IgM ("precipitin") (test may be termed “IDTP”), or IgG (CF) (test may be termed “IDCF”) antibody in body fluids. This is appropriate if no diagnosis of coccidioidomycosis has previously been established. [Non-specific reactions resembling that given by the coccidioidal precipitin (IgM) are encountered infrequently with some sera, e.g. from patients who do not have coccidioidomycosis.] 2. Quantitative: For veterinary specimens, we estimate the titer of the CF (IgG) by quantitative immunodiffusion rather than by complement fixation (CF). This provides an approximation of the CF titer. The quantitative immunodiffusion rather than the CF test avoids the problem of anticomplementary activity frequently encountered especially with canine sera. Determination of titer is appropriate after a diagnosis has been established, e.g. by positive immunodiffusion or recovery of C. immitis from the patient. B. Results and interpretation: 1. Detection of coccidioidal antibody in a single specimen can be diagnostically significant; testing sequential samples can be helpful in following the course of the disease. 2. Immunodiffusion (ID) (qualitative) - presence or not of precipitin (IgM) or CF (IgG) antibody. The IgM is important in the diagnosis of acute primary coccidioidomycosis, and is detectable in most patients in 1 to 2 weeks after onset of symptoms, may persist several months, occasionally longer in association with a pulmonary cavity or disseminated disease. -

I M M U N O L O G Y Core Notes
II MM MM UU NN OO LL OO GG YY CCOORREE NNOOTTEESS MEDICAL IMMUNOLOGY 544 FALL 2011 Dr. George A. Gutman SCHOOL OF MEDICINE UNIVERSITY OF CALIFORNIA, IRVINE (Copyright) 2011 Regents of the University of California TABLE OF CONTENTS CHAPTER 1 INTRODUCTION...................................................................................... 3 CHAPTER 2 ANTIGEN/ANTIBODY INTERACTIONS ..............................................9 CHAPTER 3 ANTIBODY STRUCTURE I..................................................................17 CHAPTER 4 ANTIBODY STRUCTURE II.................................................................23 CHAPTER 5 COMPLEMENT...................................................................................... 33 CHAPTER 6 ANTIBODY GENETICS, ISOTYPES, ALLOTYPES, IDIOTYPES.....45 CHAPTER 7 CELLULAR BASIS OF ANTIBODY DIVERSITY: CLONAL SELECTION..................................................................53 CHAPTER 8 GENETIC BASIS OF ANTIBODY DIVERSITY...................................61 CHAPTER 9 IMMUNOGLOBULIN BIOSYNTHESIS ...............................................69 CHAPTER 10 BLOOD GROUPS: ABO AND Rh .........................................................77 CHAPTER 11 CELL-MEDIATED IMMUNITY AND MHC ........................................83 CHAPTER 12 CELL INTERACTIONS IN CELL MEDIATED IMMUNITY ..............91 CHAPTER 13 T-CELL/B-CELL COOPERATION IN HUMORAL IMMUNITY......105 CHAPTER 14 CELL SURFACE MARKERS OF T-CELLS, B-CELLS AND MACROPHAGES...............................................................111 -

Practical Write up B.Sc Life Sciences Sem Vi Paper: Immunology Teachers
Practical write up B.Sc Life Sciences Sem Vi Paper: Immunology Teachers: Dr. Anita Gulati, Dr. Renu Solanki Immunology Page 1 Aim: To perform a simple immune diffusion test (Ouchterlony test) Requirements: 1.5% Agar, 1% Agar, Acetocarmine, Methylene blue, Glass slides, Micropiettes, Microtips, Blotting sheets, Tissue rolls, Plastic trays Theory: The interaction between an antibody and a soluble antigen in aqueous solution forms a lattice that eventually develops into a visible precipitate. Excess of either antibody or antigen interferes with maximal precipitation, which occurs in the so called equivalence zone, when the ratio of antibody to antigen is optimal. Figure 1: A precipitation curve for a system of one antigen and its antibodies. Immunology Page 2 Immune precipitates can form not only in solution but also in agar matrix. When antigen and antibody diffuse toward one another in agar, or when antibody is incorporated into the agar and antigen diffuses into the antibody containing matrix, a visible line of precipitation will form. Two immune diffusion techniques are radial immunodiffusion (Mancini method) and double immunodiffusion (Ouchterlony method); both are carried out in a semisolid medium such as agar. 1. Radial Immunodiffusion (Mancini Method): The relative concentrations of an antigen can be determined by a simple quantitative assay in which an antigen sample is placed in a well and allowed to diffuse into agar containing a suitable dilution of antiserum. As the antigen diffuses into the agar, the region of equivalence is established and a ring of precipitation, a precipitin ring, forms around the well. The area of precipitin ring is proportional to the concentration of antigen. -
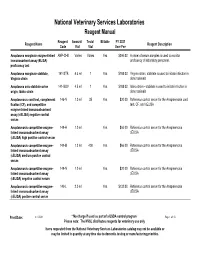
NVSL Reagent Manual
National Veterinary Services Laboratories Reagent Manual Reagent Amount/ Tests/ Billable FY 2021 Reagent Name Reagent Description Code Vial Vial User Fee Anaplasma marginale enzyme-linked ANP-CHK Varies VariesYes $394.00 A panel of serum samples is used to monitor immunosorbent assay (ELISA) proficiency of laboratory personnel. proficiency test Anaplasma marginale stabilate, 141-STB 4.5 ml 1Yes $188.00 Virginia strain, stabilate is used to initiate infection in Virginia strain donor animals Anaplasma ovis stabilate ovine 141-SOV 4.5 ml 1Yes $188.00 Idaho strain - stabilate is used to initate infection in origin, Idaho strain donor animals Anaplasmosis card test, complement 146-N 1.0 ml 35Yes $20.00 Reference control serum for the Anaplasmosis card fixation (CF), and competitive test, CF, and cELISA enzyme-linked immunoabsorbent assay (cELISA) negative control serum Anaplasmosis competitive enzyme- 149-H 1.0 mlYes $66.00 Reference control serum for the Anaplasmosis linked immunoabsorbent assay cELISA (cELISA) high positive control serum Anaplasmosis competitive enzyme- 149-M 1.0 ml 400Yes $66.00 Reference control serum for the Anaplasmosis linked immunoabsorbent assay cELISA (cELISA) medium positve control serum Anaplasmosis competitive enzyme- 149-N 1.0 mlYes $20.00 Reference control serum for the Anaplasmosis linked immunoabsorbent assay cELISA (cELISA) negative control serum Anaplasmosis competitive enzyme- 149-L 2.0 mlYes $132.00 Reference control serum for the Anaplasmosis linked immunoabsorbent assay cELISA (cELISA) positve control serum Print Date: 6/11/2021 * No charge if used as part of a USDA control program Page 1 of 58 Please note: The NVSL distributes reagents for veterinary use only Items requested from the National Veterinary Services Laboratories catalog may not be available or may be limited in quantity at any time due to domestic testing or manufacturing priorities.