IFN Signaling and Neutrophil Degranulation Transcriptional Signatures Are Induced During
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Gene Symbol Category ACAN ECM ADAM10 ECM Remodeling-Related ADAM11 ECM Remodeling-Related ADAM12 ECM Remodeling-Related ADAM15 E
Supplementary Material (ESI) for Integrative Biology This journal is (c) The Royal Society of Chemistry 2010 Gene symbol Category ACAN ECM ADAM10 ECM remodeling-related ADAM11 ECM remodeling-related ADAM12 ECM remodeling-related ADAM15 ECM remodeling-related ADAM17 ECM remodeling-related ADAM18 ECM remodeling-related ADAM19 ECM remodeling-related ADAM2 ECM remodeling-related ADAM20 ECM remodeling-related ADAM21 ECM remodeling-related ADAM22 ECM remodeling-related ADAM23 ECM remodeling-related ADAM28 ECM remodeling-related ADAM29 ECM remodeling-related ADAM3 ECM remodeling-related ADAM30 ECM remodeling-related ADAM5 ECM remodeling-related ADAM7 ECM remodeling-related ADAM8 ECM remodeling-related ADAM9 ECM remodeling-related ADAMTS1 ECM remodeling-related ADAMTS10 ECM remodeling-related ADAMTS12 ECM remodeling-related ADAMTS13 ECM remodeling-related ADAMTS14 ECM remodeling-related ADAMTS15 ECM remodeling-related ADAMTS16 ECM remodeling-related ADAMTS17 ECM remodeling-related ADAMTS18 ECM remodeling-related ADAMTS19 ECM remodeling-related ADAMTS2 ECM remodeling-related ADAMTS20 ECM remodeling-related ADAMTS3 ECM remodeling-related ADAMTS4 ECM remodeling-related ADAMTS5 ECM remodeling-related ADAMTS6 ECM remodeling-related ADAMTS7 ECM remodeling-related ADAMTS8 ECM remodeling-related ADAMTS9 ECM remodeling-related ADAMTSL1 ECM remodeling-related ADAMTSL2 ECM remodeling-related ADAMTSL3 ECM remodeling-related ADAMTSL4 ECM remodeling-related ADAMTSL5 ECM remodeling-related AGRIN ECM ALCAM Cell-cell adhesion ANGPT1 Soluble factors and receptors -

Effect of Nanoparticles on the Expression and Activity of Matrix Metalloproteinases
Nanotechnol Rev 2018; 7(6): 541–553 Review Magdalena Matysiak-Kucharek*, Magdalena Czajka, Krzysztof Sawicki, Marcin Kruszewski and Lucyna Kapka-Skrzypczak Effect of nanoparticles on the expression and activity of matrix metalloproteinases https://doi.org/10.1515/ntrev-2018-0110 Received September 14, 2018; accepted October 11, 2018; previously 1 Introduction published online November 15, 2018 Matrix metallopeptidases, commonly known as matrix Abstract: Matrix metallopeptidases, commonly known metalloproteinases (MMPs), are zinc-dependent proteo- as matrix metalloproteinases (MMPs), are a group of pro- lytic enzymes whose primary function is the degradation teolytic enzymes whose main function is the remodeling and remodeling of extracellular matrix (ECM) compo- of the extracellular matrix. Changes in the activity of nents. ECM is a complex, dynamic structure that condi- these enzymes are observed in many pathological states, tions the proper tissue architecture. MMPs by digesting including cancer metastases. An increasing body of evi- ECM proteins eliminate structural barriers and allow dence indicates that nanoparticles (NPs) can lead to the cell migration. Moreover, by hydrolyzing extracellularly deregulation of MMP expression and/or activity both in released proteins, MMPs can change the activity of many vitro and in vivo. In this work, we summarized the current signal peptides, such as growth factors, cytokines, and state of knowledge on the impact of NPs on MMPs. The chemokines. MMPs are involved in many physiological literature analysis showed that the impact of NPs on MMP processes, such as embryogenesis, reproduction cycle, or expression and/or activity is inconclusive. NPs exhibit wound healing; however, their increased activity is also both stimulating and inhibitory effects, which might be associated with a number of pathological conditions, such dependent on multiple factors, such as NP size and coat- as diabetes, cardiovascular diseases and neurodegenera- ing or a cellular model used in the research. -
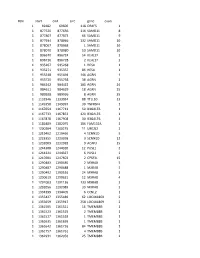
Chr Start End Size Gene Exon 1 69482 69600 118 OR4F5 1 1 877520
#chr start end size gene exon 1 69482 69600 118 OR4F5 1 1 877520 877636 116 SAMD11 8 1 877807 877873 66 SAMD11 9 1 877934 878066 132 SAMD11 10 1 878067 878068 1 SAMD11 10 1 878070 878080 10 SAMD11 10 1 896670 896724 54 KLHL17 2 1 896726 896728 2 KLHL17 2 1 935267 935268 1 HES4 1 1 935271 935357 86 HES4 1 1 955548 955694 146 AGRN 1 1 955720 955758 38 AGRN 1 1 984242 984422 180 AGRN 24 1 984611 984629 18 AGRN 25 1 989928 989936 8 AGRN 35 1 1132946 1133034 88 TTLL10 13 1 1149358 1149397 39 TNFRSF4 1 1 1167654 1167713 59 B3GALT6 1 1 1167733 1167853 120 B3GALT6 1 1 1167878 1167908 30 B3GALT6 1 1 1181889 1182075 186 FAM132A 1 1 1200204 1200215 11 UBE2J2 2 1 1219462 1219466 4 SCNN1D 5 1 1223355 1223358 3 SCNN1D 12 1 1232009 1232018 9 ACAP3 15 1 1244308 1244320 12 PUSL1 2 1 1244321 1244327 6 PUSL1 2 1 1247601 1247603 2 CPSF3L 15 1 1290483 1290485 2 MXRA8 5 1 1290487 1290488 1 MXRA8 5 1 1290492 1290516 24 MXRA8 5 1 1290619 1290631 12 MXRA8 4 1 1291003 1291136 133 MXRA8 3 1 1292056 1292089 33 MXRA8 2 1 1334399 1334405 6 CCNL2 1 1 1355427 1355489 62 LOC441869 2 1 1355659 1355917 258 LOC441869 2 1 1361505 1361521 16 TMEM88B 1 1 1361523 1361525 2 TMEM88B 1 1 1361527 1361528 1 TMEM88B 1 1 1361635 1361636 1 TMEM88B 1 1 1361642 1361726 84 TMEM88B 1 1 1361757 1361761 4 TMEM88B 1 1 1362931 1362956 25 TMEM88B 2 1 1374780 1374838 58 VWA1 3 1 1374841 1374842 1 VWA1 3 1 1374934 1375100 166 VWA1 3 1 1389738 1389747 9 ATAD3C 4 1 1389751 1389816 65 ATAD3C 4 1 1389830 1389849 19 ATAD3C 4 1 1390835 1390837 2 ATAD3C 5 1 1407260 1407331 71 ATAD3B 1 1 1407340 1407474 -

(12) United States Patent (10) Patent No.: US 7.873,482 B2 Stefanon Et Al
US007873482B2 (12) United States Patent (10) Patent No.: US 7.873,482 B2 Stefanon et al. (45) Date of Patent: Jan. 18, 2011 (54) DIAGNOSTIC SYSTEM FOR SELECTING 6,358,546 B1 3/2002 Bebiak et al. NUTRITION AND PHARMACOLOGICAL 6,493,641 B1 12/2002 Singh et al. PRODUCTS FOR ANIMALS 6,537,213 B2 3/2003 Dodds (76) Inventors: Bruno Stefanon, via Zilli, 51/A/3, Martignacco (IT) 33035: W. Jean Dodds, 938 Stanford St., Santa Monica, (Continued) CA (US) 90403 FOREIGN PATENT DOCUMENTS (*) Notice: Subject to any disclaimer, the term of this patent is extended or adjusted under 35 WO WO99-67642 A2 12/1999 U.S.C. 154(b) by 158 days. (21)21) Appl. NoNo.: 12/316,8249 (Continued) (65) Prior Publication Data Swanson, et al., “Nutritional Genomics: Implication for Companion Animals'. The American Society for Nutritional Sciences, (2003).J. US 2010/O15301.6 A1 Jun. 17, 2010 Nutr. 133:3033-3040 (18 pages). (51) Int. Cl. (Continued) G06F 9/00 (2006.01) (52) U.S. Cl. ........................................................ 702/19 Primary Examiner—Edward Raymond (58) Field of Classification Search ................... 702/19 (74) Attorney, Agent, or Firm Greenberg Traurig, LLP 702/23, 182–185 See application file for complete search history. (57) ABSTRACT (56) References Cited An analysis of the profile of a non-human animal comprises: U.S. PATENT DOCUMENTS a) providing a genotypic database to the species of the non 3,995,019 A 1 1/1976 Jerome human animal Subject or a selected group of the species; b) 5,691,157 A 1 1/1997 Gong et al. -

Computational Analyses of Small Molecules Activity from Phenotypic Screens
Computational analyses of small molecules activity from phenotypic screens Azedine Zoufir Hughes Hall This dissertation is submitted for the degree of Doctor of Philosophy July 2018 Declaration This thesis is submitted as the result of my own work and includes nothing which is the outcome of work done in collaboration except where specifically indicated in the text. It is not substantially the same as any that I have submitted, or, is being concurrently submitted for a degree or diploma or other qualification at the University of Cambridge or any other University or similar institution except as declared in the preface and specified in the text. I further state that no substantial part of my dissertation has already been submitted, or, is being concurrently submitted for any such degree, diploma or other qualification at the University of Cambridge or any other University or similar institution except as declared in the Preface and specified in the text. This dissertation does not exceed the word limit of 60,000 words. Azedine Zoufir July 2018 Summary Title: Computational analyses of small molecules activity from phenotypic screens Author: Azedine Zoufir Drug discovery is no longer relying on the one gene-one disease paradigm nor on target-based screening alone to discover new drugs. Phenotypic-based screening is regaining momentum to discover new compounds since those assays provide an environment closer to the physiological state of the disease and allow to better anticipate off-target effects and other factors that can limit the efficacy of the drugs. However, uncovering the mechanism of action of the compounds active in those assays relies on in vitro techniques that are expensive and time- consuming. -

Human Induced Pluripotent Stem Cell–Derived Podocytes Mature Into Vascularized Glomeruli Upon Experimental Transplantation
BASIC RESEARCH www.jasn.org Human Induced Pluripotent Stem Cell–Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation † Sazia Sharmin,* Atsuhiro Taguchi,* Yusuke Kaku,* Yasuhiro Yoshimura,* Tomoko Ohmori,* ‡ † ‡ Tetsushi Sakuma, Masashi Mukoyama, Takashi Yamamoto, Hidetake Kurihara,§ and | Ryuichi Nishinakamura* *Department of Kidney Development, Institute of Molecular Embryology and Genetics, and †Department of Nephrology, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan; ‡Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University, Hiroshima, Japan; §Division of Anatomy, Juntendo University School of Medicine, Tokyo, Japan; and |Japan Science and Technology Agency, CREST, Kumamoto, Japan ABSTRACT Glomerular podocytes express proteins, such as nephrin, that constitute the slit diaphragm, thereby contributing to the filtration process in the kidney. Glomerular development has been analyzed mainly in mice, whereas analysis of human kidney development has been minimal because of limited access to embryonic kidneys. We previously reported the induction of three-dimensional primordial glomeruli from human induced pluripotent stem (iPS) cells. Here, using transcription activator–like effector nuclease-mediated homologous recombination, we generated human iPS cell lines that express green fluorescent protein (GFP) in the NPHS1 locus, which encodes nephrin, and we show that GFP expression facilitated accurate visualization of nephrin-positive podocyte formation in -

Single Cell Derived Clonal Analysis of Human Glioblastoma Links
SUPPLEMENTARY INFORMATION: Single cell derived clonal analysis of human glioblastoma links functional and genomic heterogeneity ! Mona Meyer*, Jüri Reimand*, Xiaoyang Lan, Renee Head, Xueming Zhu, Michelle Kushida, Jane Bayani, Jessica C. Pressey, Anath Lionel, Ian D. Clarke, Michael Cusimano, Jeremy Squire, Stephen Scherer, Mark Bernstein, Melanie A. Woodin, Gary D. Bader**, and Peter B. Dirks**! ! * These authors contributed equally to this work.! ** Correspondence: [email protected] or [email protected]! ! Supplementary information - Meyer, Reimand et al. Supplementary methods" 4" Patient samples and fluorescence activated cell sorting (FACS)! 4! Differentiation! 4! Immunocytochemistry and EdU Imaging! 4! Proliferation! 5! Western blotting ! 5! Temozolomide treatment! 5! NCI drug library screen! 6! Orthotopic injections! 6! Immunohistochemistry on tumor sections! 6! Promoter methylation of MGMT! 6! Fluorescence in situ Hybridization (FISH)! 7! SNP6 microarray analysis and genome segmentation! 7! Calling copy number alterations! 8! Mapping altered genome segments to genes! 8! Recurrently altered genes with clonal variability! 9! Global analyses of copy number alterations! 9! Phylogenetic analysis of copy number alterations! 10! Microarray analysis! 10! Gene expression differences of TMZ resistant and sensitive clones of GBM-482! 10! Reverse transcription-PCR analyses! 11! Tumor subtype analysis of TMZ-sensitive and resistant clones! 11! Pathway analysis of gene expression in the TMZ-sensitive clone of GBM-482! 11! Supplementary figures and tables" 13" "2 Supplementary information - Meyer, Reimand et al. Table S1: Individual clones from all patient tumors are tumorigenic. ! 14! Fig. S1: clonal tumorigenicity.! 15! Fig. S2: clonal heterogeneity of EGFR and PTEN expression.! 20! Fig. S3: clonal heterogeneity of proliferation.! 21! Fig. -

Matrisome-Associated Gene Expression Patterns Correlating with TIMP2 in Cancer David Peeney1*, Yu Fan2, Trinh Nguyen 2, Daoud Meerzaman2 & William G
www.nature.com/scientificreports OPEN Matrisome-Associated Gene Expression Patterns Correlating with TIMP2 in Cancer David Peeney1*, Yu Fan2, Trinh Nguyen 2, Daoud Meerzaman2 & William G. Stetler-Stevenson 1 Remodeling of the extracellular matrix (ECM) to facilitate invasion and metastasis is a universal hallmark of cancer progression. However, a defnitive therapeutic target remains to be identifed in this tissue compartment. As major modulators of ECM structure and function, matrix metalloproteinases (MMPs) are highly expressed in cancer and have been shown to support tumor progression. MMP enzymatic activity is inhibited by the tissue inhibitor of metalloproteinase (TIMP1–4) family of proteins, suggesting that TIMPs may possess anti-tumor activity. TIMP2 is a promiscuous MMP inhibitor that is ubiquitously expressed in normal tissues. In this study, we address inconsistencies in the literature regarding the role of TIMP2 in tumor progression by analyzing co-expressed genes in tumor vs. normal tissue. Utilizing data from The Cancer Genome Atlas and Genotype-Tissue expression studies, focusing on breast and lung carcinomas, we analyzed the correlation between TIMP2 expression and the transcriptome to identify a list of genes whose expression is highly correlated with TIMP2 in tumor tissues. Bioinformatic analysis of the identifed gene list highlights a core of matrix and matrix- associated genes that are of interest as potential modulators of TIMP2 function, thus ECM structure, identifying potential tumor microenvironment biomarkers and/or therapeutic targets for further study. Matrix metalloproteinases (MMPs) are a family of zinc-dependent endopeptidases that are the major mediators of extracellular matrix (ECM) breakdown and turnover. Humans express 23 MMPs and these play a critical role in tissue development, homeostasis and disease progression. -

A Pan-Cancer Perspective of Matrix Metalloproteases (MMP) Gene
Gobin et al. BMC Cancer (2019) 19:581 https://doi.org/10.1186/s12885-019-5768-0 RESEARCHARTICLE Open Access A pan-cancer perspective of matrix metalloproteases (MMP) gene expression profile and their diagnostic/prognostic potential Emily Gobin†, Kayla Bagwell†, John Wagner†, David Mysona, Sharmila Sandirasegarane, Nathan Smith, Shan Bai, Ashok Sharma, Robert Schleifer and Jin-Xiong She* Abstract Implication: By understanding Matrix Metalloprotease (MMP) dysregulation from a pan-cancer perspective, this study sheds light on the diagnostic potentials of MMPs across multiple neoplasms. Background: MMPs are intriguing genes related to cancer disease progression, functional promotion of angiogenesis, invasion, metastasis, and avoidance of immune surveillance. Many studies have noted these genes are frequently upregulated in cancer. However, expression patterns of all MMPs and their diagnostic and prognostic potential have not been investigated in a pan-cancer perspective. Methods: The Cancer Genome Atlas (TCGA) data were used to evaluate diagnostic and prognostic potential of 24 MMPs in fifteen different cancer types. Gene expression measured by RNA-seq was analyzed by differential expression, hierarchical clustering, and ROC analysis for individual genes and in combination. Results: MMP1, MMP9, MMP10, MMP11, and MMP13 were almost universally upregulated across all cancers, with significant (p < 0.05) fold change (FC > 2) in ten of fifteen cancers. MMP3, MMP7, MMP12 and MMP14) are significantly up-regulated in at least 10 cancer types. Interestingly, MMP2, MMP7, MMP23B, MMP27 and MMP28) are significantly down-regulated in seven to nine cancer types. Multiple MMPs possess AUC’s > 0.9 in more than one cancer. However, survival analyses suggest that the prognostic value of MMPs is limited to clear cell renal carcinoma. -

A Genomic Analysis of Rat Proteases and Protease Inhibitors
A genomic analysis of rat proteases and protease inhibitors Xose S. Puente and Carlos López-Otín Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, 33006-Oviedo, Spain Send correspondence to: Carlos López-Otín Departamento de Bioquímica y Biología Molecular Facultad de Medicina, Universidad de Oviedo 33006 Oviedo-SPAIN Tel. 34-985-104201; Fax: 34-985-103564 E-mail: [email protected] Proteases perform fundamental roles in multiple biological processes and are associated with a growing number of pathological conditions that involve abnormal or deficient functions of these enzymes. The availability of the rat genome sequence has opened the possibility to perform a global analysis of the complete protease repertoire or degradome of this model organism. The rat degradome consists of at least 626 proteases and homologs, which are distributed into five catalytic classes: 24 aspartic, 160 cysteine, 192 metallo, 221 serine, and 29 threonine proteases. Overall, this distribution is similar to that of the mouse degradome, but significatively more complex than that corresponding to the human degradome composed of 561 proteases and homologs. This increased complexity of the rat protease complement mainly derives from the expansion of several gene families including placental cathepsins, testases, kallikreins and hematopoietic serine proteases, involved in reproductive or immunological functions. These protease families have also evolved differently in the rat and mouse genomes and may contribute to explain some functional differences between these two closely related species. Likewise, genomic analysis of rat protease inhibitors has shown some differences with the mouse protease inhibitor complement and the marked expansion of families of cysteine and serine protease inhibitors in rat and mouse with respect to human. -

168 1. ABSTRACT 2. INTRODUCTION Matrix Metalloproteinase Function
[Frontiers in Bioscience, Scholar, 7, 168-183, June 1, 2015] Matrix metalloproteinase function in non-mammalian model organisms Julia J. Buckley1, Jason R. Jessen1 1Department of Biology, Middle Tennessee State University, Murfreesboro, TN 37130 TABLE OF CONTENTS 1. Abstract 2. Introduction 3. Flowering Plants 4. Hydra 5. Amphioxus 6. Sea Urchin 7. Worm 8. Fly 9. Frog 10. Zebrafish 11. Chicken 12. Conclusions 13. Acknowledgements 14. References 1. ABSTRACT Matrix metalloproteinases (MMPs), members of the metzincin superfamily of zinc-dependent adamalysins, astacins, and serralysins are members of endopeptidases (2). MMPs are capable of cleaving the metzincin superfamily of proteases. MMPs constitute extracellular matrix (ECM) and non-ECM proteins a large protein family of both secreted and membrane- including adhesion molecules, ligands, and receptors. tethered enzymes that are synthesized as zymogens The human genome contains 23 MMP genes and 4 (proMMP) and activated by a cysteine-switch mechanism. genes encoding tissue inhibitors of metalloproteinases First described over 50 years ago by Gross and Lapiere (TIMPs). Most human MMP proteins have a similar as a collagenolytic activity in amphibian tissues, the domain organization that includes a signal peptide, human MMP family now encompasses 23 different propeptide domain, catalytic domain, hinge region, and genes whose encoded proteins are capable of cleaving a hemopexin domain. While many MMPs are secreted a variety of extracellular matrix protein substrates. Since as inactive zymogens requiring proprotein convertase their expression is upregulated in many cancer cell types, activity to remove the N-terminal propeptide domain for MMPs have received much attention particularly in the activation (3), six MMPs are membrane-tethered and are areas of tumor progression and metastasis. -

Matrix Metalloproteinases As Potential Biomarkers and Therapeutic Targets in Liver Diseases
cells Review Matrix Metalloproteinases as Potential Biomarkers and Therapeutic Targets in Liver Diseases Eline Geervliet and Ruchi Bansal * Translational Liver Research, Department of Medical Cell BioPhysics, Technical Medical Centre, Faculty of Science and Technology, University of Twente, 7522 NB Enschede, The Netherlands; [email protected] * Correspondence: [email protected]; Tel.: +31-53-489-3115 Received: 6 April 2020; Accepted: 13 May 2020; Published: 13 May 2020 Abstract: Chronic liver diseases, characterized by an excessive accumulation of extracellular matrix (ECM) resulting in scar tissue formation, are a growing health problem causing increasing morbidity and mortality worldwide. Currently, therapeutic options for tissue fibrosis are severely limited, and organ transplantation is the only treatment for the end-stage liver diseases. During liver damage, injured hepatocytes release proinflammatory factors resulting in the recruitment and activation of immune cells that activate quiescent hepatic stellate cells (HSCs). Upon activation, HSCs transdifferentiate into highly proliferative, migratory, contractile and ECM-producing myofibroblasts. The disrupted balance between ECM deposition and degradation leads to the formation of scar tissue referred to as fibrosis. This balance can be restored either by reducing ECM deposition (by inhibition of HSCs activation and proliferation) or enhancing ECM degradation (by increased expression of matrix metalloproteinases (MMPs)). MMPs play an important role in ECM remodeling and represent an interesting target for therapeutic drug discovery. In this review, we present the current knowledge about ECM remodeling and role of the different MMPs in liver diseases. MMP expression patterns in different stages of liver diseases have also been reviewed to determine their role as biomarkers.