A Promising Target to Limit the Local Invasiveness of Colorectal Cancer
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ASPH-Notch Axis Guided Exosomal Delivery of Prometastatic Secretome
Lin et al. Molecular Cancer (2019) 18:156 https://doi.org/10.1186/s12943-019-1077-0 RESEARCH Open Access ASPH-notch Axis guided Exosomal delivery of Prometastatic Secretome renders breast Cancer multi-organ metastasis Qiushi Lin1†, Xuesong Chen2†, Fanzheng Meng3†, Kosuke Ogawa4†, Min Li5, Ruipeng Song3, Shugeng Zhang3, Ziran Zhang3, Xianglu Kong3, Qinggang Xu1,6, Fuliang He1,7, Xuewei Bai8, Bei Sun8, Mien-Chie Hung9,10, Lianxin Liu3,11*, Jack Wands4* and Xiaoqun Dong12,1* Abstract Background: Aspartate β-hydroxylase (ASPH) is silent in normal adult tissues only to re-emerge during oncogenesis where its function is required for generation and maintenance of malignant phenotypes. Exosomes enable prooncogenic secretome delivering and trafficking for long-distance cell-to-cell communication. This study aims to explore molecular mechanisms underlying how ASPH network regulates designated exosomes to program development and progression of breast cancer. Methods: Stable cell lines overexpressing or knocking-out of ASPH were established using lentivirus transfection or CRISPR-CAS9 systems. Western blot, MTT, immunofluorescence, luciferase reporter, co-immunoprecipitation, 2D/3-D invasion, tube formation, mammosphere formation, immunohistochemistry and newly developed in vitro metastasis were applied. Results: Through physical interactions with Notch receptors, ligands (JAGs) and regulators (ADAM10/17), ASPH activates Notch cascade to provide raw materials (especially MMPs/ADAMs) for synthesis/release of pro-metastatic exosomes. Exosomes orchestrate EMT, 2-D/3-D invasion, stemness, angiogenesis, and premetastatic niche formation. Small molecule inhibitors (SMIs) of ASPH’s β-hydroxylase specifically/efficiently abrogated in vitro metastasis, which mimics basement membrane invasion at primary site, intravasation/extravasation (transendothelial migration), and colonization/outgrowth at distant sites. -

Mutant IDH, (R)-2-Hydroxyglutarate, and Cancer
Downloaded from genesdev.cshlp.org on October 1, 2021 - Published by Cold Spring Harbor Laboratory Press REVIEW What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer Julie-Aurore Losman1 and William G. Kaelin Jr.1,2,3 1Department of Medical Oncology, Dana-Farber Cancer Institute, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts 02215, USA; 2Howard Hughes Medical Institute, Chevy Chase, Maryland 20815, USA Mutations in metabolic enzymes, including isocitrate whether altered cellular metabolism is a cause of cancer dehydrogenase 1 (IDH1) and IDH2, in cancer strongly or merely an adaptive response of cancer cells in the face implicate altered metabolism in tumorigenesis. IDH1 of accelerated cell proliferation is still a topic of some and IDH2 catalyze the interconversion of isocitrate and debate. 2-oxoglutarate (2OG). 2OG is a TCA cycle intermediate The recent identification of cancer-associated muta- and an essential cofactor for many enzymes, including tions in three metabolic enzymes suggests that altered JmjC domain-containing histone demethylases, TET cellular metabolism can indeed be a cause of some 5-methylcytosine hydroxylases, and EglN prolyl-4-hydrox- cancers (Pollard et al. 2003; King et al. 2006; Raimundo ylases. Cancer-associated IDH mutations alter the enzymes et al. 2011). Two of these enzymes, fumarate hydratase such that they reduce 2OG to the structurally similar (FH) and succinate dehydrogenase (SDH), are bone fide metabolite (R)-2-hydroxyglutarate [(R)-2HG]. Here we tumor suppressors, and loss-of-function mutations in FH review what is known about the molecular mechanisms and SDH have been identified in various cancers, in- of transformation by mutant IDH and discuss their im- cluding renal cell carcinomas and paragangliomas. -

Gene Symbol Category ACAN ECM ADAM10 ECM Remodeling-Related ADAM11 ECM Remodeling-Related ADAM12 ECM Remodeling-Related ADAM15 E
Supplementary Material (ESI) for Integrative Biology This journal is (c) The Royal Society of Chemistry 2010 Gene symbol Category ACAN ECM ADAM10 ECM remodeling-related ADAM11 ECM remodeling-related ADAM12 ECM remodeling-related ADAM15 ECM remodeling-related ADAM17 ECM remodeling-related ADAM18 ECM remodeling-related ADAM19 ECM remodeling-related ADAM2 ECM remodeling-related ADAM20 ECM remodeling-related ADAM21 ECM remodeling-related ADAM22 ECM remodeling-related ADAM23 ECM remodeling-related ADAM28 ECM remodeling-related ADAM29 ECM remodeling-related ADAM3 ECM remodeling-related ADAM30 ECM remodeling-related ADAM5 ECM remodeling-related ADAM7 ECM remodeling-related ADAM8 ECM remodeling-related ADAM9 ECM remodeling-related ADAMTS1 ECM remodeling-related ADAMTS10 ECM remodeling-related ADAMTS12 ECM remodeling-related ADAMTS13 ECM remodeling-related ADAMTS14 ECM remodeling-related ADAMTS15 ECM remodeling-related ADAMTS16 ECM remodeling-related ADAMTS17 ECM remodeling-related ADAMTS18 ECM remodeling-related ADAMTS19 ECM remodeling-related ADAMTS2 ECM remodeling-related ADAMTS20 ECM remodeling-related ADAMTS3 ECM remodeling-related ADAMTS4 ECM remodeling-related ADAMTS5 ECM remodeling-related ADAMTS6 ECM remodeling-related ADAMTS7 ECM remodeling-related ADAMTS8 ECM remodeling-related ADAMTS9 ECM remodeling-related ADAMTSL1 ECM remodeling-related ADAMTSL2 ECM remodeling-related ADAMTSL3 ECM remodeling-related ADAMTSL4 ECM remodeling-related ADAMTSL5 ECM remodeling-related AGRIN ECM ALCAM Cell-cell adhesion ANGPT1 Soluble factors and receptors -

Effect of Nanoparticles on the Expression and Activity of Matrix Metalloproteinases
Nanotechnol Rev 2018; 7(6): 541–553 Review Magdalena Matysiak-Kucharek*, Magdalena Czajka, Krzysztof Sawicki, Marcin Kruszewski and Lucyna Kapka-Skrzypczak Effect of nanoparticles on the expression and activity of matrix metalloproteinases https://doi.org/10.1515/ntrev-2018-0110 Received September 14, 2018; accepted October 11, 2018; previously 1 Introduction published online November 15, 2018 Matrix metallopeptidases, commonly known as matrix Abstract: Matrix metallopeptidases, commonly known metalloproteinases (MMPs), are zinc-dependent proteo- as matrix metalloproteinases (MMPs), are a group of pro- lytic enzymes whose primary function is the degradation teolytic enzymes whose main function is the remodeling and remodeling of extracellular matrix (ECM) compo- of the extracellular matrix. Changes in the activity of nents. ECM is a complex, dynamic structure that condi- these enzymes are observed in many pathological states, tions the proper tissue architecture. MMPs by digesting including cancer metastases. An increasing body of evi- ECM proteins eliminate structural barriers and allow dence indicates that nanoparticles (NPs) can lead to the cell migration. Moreover, by hydrolyzing extracellularly deregulation of MMP expression and/or activity both in released proteins, MMPs can change the activity of many vitro and in vivo. In this work, we summarized the current signal peptides, such as growth factors, cytokines, and state of knowledge on the impact of NPs on MMPs. The chemokines. MMPs are involved in many physiological literature analysis showed that the impact of NPs on MMP processes, such as embryogenesis, reproduction cycle, or expression and/or activity is inconclusive. NPs exhibit wound healing; however, their increased activity is also both stimulating and inhibitory effects, which might be associated with a number of pathological conditions, such dependent on multiple factors, such as NP size and coat- as diabetes, cardiovascular diseases and neurodegenera- ing or a cellular model used in the research. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -
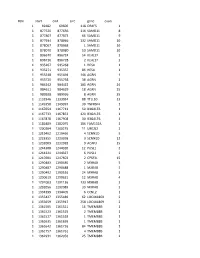
Chr Start End Size Gene Exon 1 69482 69600 118 OR4F5 1 1 877520
#chr start end size gene exon 1 69482 69600 118 OR4F5 1 1 877520 877636 116 SAMD11 8 1 877807 877873 66 SAMD11 9 1 877934 878066 132 SAMD11 10 1 878067 878068 1 SAMD11 10 1 878070 878080 10 SAMD11 10 1 896670 896724 54 KLHL17 2 1 896726 896728 2 KLHL17 2 1 935267 935268 1 HES4 1 1 935271 935357 86 HES4 1 1 955548 955694 146 AGRN 1 1 955720 955758 38 AGRN 1 1 984242 984422 180 AGRN 24 1 984611 984629 18 AGRN 25 1 989928 989936 8 AGRN 35 1 1132946 1133034 88 TTLL10 13 1 1149358 1149397 39 TNFRSF4 1 1 1167654 1167713 59 B3GALT6 1 1 1167733 1167853 120 B3GALT6 1 1 1167878 1167908 30 B3GALT6 1 1 1181889 1182075 186 FAM132A 1 1 1200204 1200215 11 UBE2J2 2 1 1219462 1219466 4 SCNN1D 5 1 1223355 1223358 3 SCNN1D 12 1 1232009 1232018 9 ACAP3 15 1 1244308 1244320 12 PUSL1 2 1 1244321 1244327 6 PUSL1 2 1 1247601 1247603 2 CPSF3L 15 1 1290483 1290485 2 MXRA8 5 1 1290487 1290488 1 MXRA8 5 1 1290492 1290516 24 MXRA8 5 1 1290619 1290631 12 MXRA8 4 1 1291003 1291136 133 MXRA8 3 1 1292056 1292089 33 MXRA8 2 1 1334399 1334405 6 CCNL2 1 1 1355427 1355489 62 LOC441869 2 1 1355659 1355917 258 LOC441869 2 1 1361505 1361521 16 TMEM88B 1 1 1361523 1361525 2 TMEM88B 1 1 1361527 1361528 1 TMEM88B 1 1 1361635 1361636 1 TMEM88B 1 1 1361642 1361726 84 TMEM88B 1 1 1361757 1361761 4 TMEM88B 1 1 1362931 1362956 25 TMEM88B 2 1 1374780 1374838 58 VWA1 3 1 1374841 1374842 1 VWA1 3 1 1374934 1375100 166 VWA1 3 1 1389738 1389747 9 ATAD3C 4 1 1389751 1389816 65 ATAD3C 4 1 1389830 1389849 19 ATAD3C 4 1 1390835 1390837 2 ATAD3C 5 1 1407260 1407331 71 ATAD3B 1 1 1407340 1407474 -

Synthesis of 2-Oxoglutarate Derivatives and Their Evaluation As Cosubstrates and Inhibitors of Human Cite This: Chem
Chemical Science View Article Online EDGE ARTICLE View Journal | View Issue Synthesis of 2-oxoglutarate derivatives and their evaluation as cosubstrates and inhibitors of human Cite this: Chem. Sci.,2021,12,1327 b † All publication charges for this article aspartate/asparagine- -hydroxylase have been paid for by the Royal Society of Chemistry Lennart Brewitz, Yu Nakashima and Christopher J. Schofield * 2-Oxoglutarate (2OG) is involved in biological processes including oxidations catalyzed by 2OG oxygenases for which it is a cosubstrate. Eukaryotic 2OG oxygenases have roles in collagen biosynthesis, lipid metabolism, DNA/RNA modification, transcriptional regulation, and the hypoxic response. Aspartate/asparagine-b-hydroxylase (AspH) is a human 2OG oxygenase catalyzing post- translational hydroxylation of Asp/Asn-residues in epidermal growth factor-like domains (EGFDs) in the endoplasmic reticulum. AspH is of chemical interest, because its Fe(II) cofactor is complexed by two rather than the typical three residues. AspH is upregulated in hypoxia and is a prognostic marker on the surface of cancer cells. We describe studies on how derivatives of its natural 2OG cosubstrate Creative Commons Attribution 3.0 Unported Licence. modulate AspH activity. An efficient synthesis of C3- and/or C4-substituted 2OG derivatives, proceeding via cyanosulfur ylid intermediates, is reported. Mass spectrometry-based AspH assays with >30 2OG derivatives reveal that some efficiently inhibit AspH via competing with 2OG as evidenced by crystallographic and solution analyses. Other 2OG derivatives can substitute for 2OG enabling substrate hydroxylation. The results show that subtle changes, e.g. methyl- to ethyl- substitution, can significantly alter the balance between catalysis and inhibition. -

Targeting the PI3K/AKT Pathway Overcomes Enzalutamide Resistance by Inhibiting
Author Manuscript Published OnlineFirst on May 5, 2020; DOI: 10.1158/1535-7163.MCT-19-0936 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. Targeting the PI3K/AKT pathway overcomes enzalutamide resistance by inhibiting induction of the glucocorticoid receptor Remi Adelaiye-Ogala1, Berkley E. Gryder2, Yen Thi Minh Nguyen1, Aian Neil Alilin1, Adlai R. Grayson1, Wardah Bajwa1, Keith H. Jansson1, Michael L. Beshiri1, Supreet Agarwal1, Jose 1 1 1 1,3* Antonio Rodriguez-Nieves , Brian Capaldo , Kathleen Kelly and David J. VanderWeele 1Laboratory for Genitourinary Cancer Pathogenesis, Center for Cancer Research, National Cancer Institute, NIH Bethesda, MD, 2Oncogenomics Section, Genetics Branch, National Cancer Institute, NIH Bethesda, MD, 3Department of Medicine, Northwestern University Feinberg School of Medicine, Chicago, IL. Running Title: Targeting the PI3K/AKT pathway and the glucocorticoid receptor Key Words: Prostate cancer, therapeutic resistance, AKT inhibition, androgen and glucocorticoid receptors Conflict of interest: The authors declare no conflict of interest. *Corresponding Author David J. VanderWeele, M.D., Ph.D. 676 N. St. Clair Suite 850 Chicago, IL . 60611 Phone: 312-926-2413 [email protected] Downloaded from mct.aacrjournals.org on October 1, 2021. © 2020 American Association for Cancer Research. Author Manuscript Published OnlineFirst on May 5, 2020; DOI: 10.1158/1535-7163.MCT-19-0936 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. ABSTRACT The PI3K-AKT pathway has pleiotropic effects, and its inhibition has long been of interest in the management of prostate cancer, where a compensatory increase in PI3K signaling has been reported following Androgen Receptor (AR) blockade. -

ASPH (F-7): Sc-365012
SAN TA C RUZ BI OTEC HNOL OG Y, INC . ASPH (F-7): sc-365012 BACKGROUND APPLICATIONS Aspartyl/asparaginyl β-hydroxylase (ASPH) is a widely-expressed type II ASPH (F-7) is recommended for detection of ASPH of mouse, rat and human membrane protein involved in calcium homeostasis. Located in the endo - origin by Western Blotting (starting dilution 1:100, dilution range 1:100- plasmic reticulum, ASPH specifically hydroxylates an Asp or Asn residue in 1:1000), immunoprecipitation [1-2 µg per 100-500 µg of total protein (1 ml the epidermal growth factor-like (EGF) domains of several proteins, using of cell lysate)], immunofluorescence (starting dilution 1:50, dilution range iron as a cofactor. The ASPH gene encodes three proteins, ASPH, Junctin and 1:50-1:500), immunohistochemistry (including paraffin-embedded sections) Junctate (or Humbug), that differ significantly in their C-terminal domains. (starting dilution 1:50, dilution range 1:50-1:500) and solid phase ELISA These ASPH gene products are expressed as five transcript variants that dif - (starting dilution 1:30, dilution range 1:30-1:3000). fer by their roles in calcium storage and release, hydroxylation capabilities Suitable for use as control antibody for ASPH siRNA (h): sc-44989, ASPH and tissue specificity. While all ASPH variants are expressed in skeletal siRNA (m): sc-44990, ASPH shRNA Plasmid (h): sc-44989-SH, ASPH shRNA muscle, only some are detected in heart, brain, pancreas, placenta, lung, Plasmid (m): sc-44990-SH, ASPH shRNA (h) Lentiviral Particles: sc-44989-V liver and kidney tissues. In the lumen of the endoplasmic reticulum, ASPH and ASPH shRNA (m) Lentiviral Particles: sc-44990-V. -

(12) United States Patent (10) Patent No.: US 7.873,482 B2 Stefanon Et Al
US007873482B2 (12) United States Patent (10) Patent No.: US 7.873,482 B2 Stefanon et al. (45) Date of Patent: Jan. 18, 2011 (54) DIAGNOSTIC SYSTEM FOR SELECTING 6,358,546 B1 3/2002 Bebiak et al. NUTRITION AND PHARMACOLOGICAL 6,493,641 B1 12/2002 Singh et al. PRODUCTS FOR ANIMALS 6,537,213 B2 3/2003 Dodds (76) Inventors: Bruno Stefanon, via Zilli, 51/A/3, Martignacco (IT) 33035: W. Jean Dodds, 938 Stanford St., Santa Monica, (Continued) CA (US) 90403 FOREIGN PATENT DOCUMENTS (*) Notice: Subject to any disclaimer, the term of this patent is extended or adjusted under 35 WO WO99-67642 A2 12/1999 U.S.C. 154(b) by 158 days. (21)21) Appl. NoNo.: 12/316,8249 (Continued) (65) Prior Publication Data Swanson, et al., “Nutritional Genomics: Implication for Companion Animals'. The American Society for Nutritional Sciences, (2003).J. US 2010/O15301.6 A1 Jun. 17, 2010 Nutr. 133:3033-3040 (18 pages). (51) Int. Cl. (Continued) G06F 9/00 (2006.01) (52) U.S. Cl. ........................................................ 702/19 Primary Examiner—Edward Raymond (58) Field of Classification Search ................... 702/19 (74) Attorney, Agent, or Firm Greenberg Traurig, LLP 702/23, 182–185 See application file for complete search history. (57) ABSTRACT (56) References Cited An analysis of the profile of a non-human animal comprises: U.S. PATENT DOCUMENTS a) providing a genotypic database to the species of the non 3,995,019 A 1 1/1976 Jerome human animal Subject or a selected group of the species; b) 5,691,157 A 1 1/1997 Gong et al. -

Computational Analyses of Small Molecules Activity from Phenotypic Screens
Computational analyses of small molecules activity from phenotypic screens Azedine Zoufir Hughes Hall This dissertation is submitted for the degree of Doctor of Philosophy July 2018 Declaration This thesis is submitted as the result of my own work and includes nothing which is the outcome of work done in collaboration except where specifically indicated in the text. It is not substantially the same as any that I have submitted, or, is being concurrently submitted for a degree or diploma or other qualification at the University of Cambridge or any other University or similar institution except as declared in the preface and specified in the text. I further state that no substantial part of my dissertation has already been submitted, or, is being concurrently submitted for any such degree, diploma or other qualification at the University of Cambridge or any other University or similar institution except as declared in the Preface and specified in the text. This dissertation does not exceed the word limit of 60,000 words. Azedine Zoufir July 2018 Summary Title: Computational analyses of small molecules activity from phenotypic screens Author: Azedine Zoufir Drug discovery is no longer relying on the one gene-one disease paradigm nor on target-based screening alone to discover new drugs. Phenotypic-based screening is regaining momentum to discover new compounds since those assays provide an environment closer to the physiological state of the disease and allow to better anticipate off-target effects and other factors that can limit the efficacy of the drugs. However, uncovering the mechanism of action of the compounds active in those assays relies on in vitro techniques that are expensive and time- consuming. -

Human Induced Pluripotent Stem Cell–Derived Podocytes Mature Into Vascularized Glomeruli Upon Experimental Transplantation
BASIC RESEARCH www.jasn.org Human Induced Pluripotent Stem Cell–Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation † Sazia Sharmin,* Atsuhiro Taguchi,* Yusuke Kaku,* Yasuhiro Yoshimura,* Tomoko Ohmori,* ‡ † ‡ Tetsushi Sakuma, Masashi Mukoyama, Takashi Yamamoto, Hidetake Kurihara,§ and | Ryuichi Nishinakamura* *Department of Kidney Development, Institute of Molecular Embryology and Genetics, and †Department of Nephrology, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan; ‡Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University, Hiroshima, Japan; §Division of Anatomy, Juntendo University School of Medicine, Tokyo, Japan; and |Japan Science and Technology Agency, CREST, Kumamoto, Japan ABSTRACT Glomerular podocytes express proteins, such as nephrin, that constitute the slit diaphragm, thereby contributing to the filtration process in the kidney. Glomerular development has been analyzed mainly in mice, whereas analysis of human kidney development has been minimal because of limited access to embryonic kidneys. We previously reported the induction of three-dimensional primordial glomeruli from human induced pluripotent stem (iPS) cells. Here, using transcription activator–like effector nuclease-mediated homologous recombination, we generated human iPS cell lines that express green fluorescent protein (GFP) in the NPHS1 locus, which encodes nephrin, and we show that GFP expression facilitated accurate visualization of nephrin-positive podocyte formation in