Matrisome-Associated Gene Expression Patterns Correlating with TIMP2 in Cancer David Peeney1*, Yu Fan2, Trinh Nguyen 2, Daoud Meerzaman2 & William G
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Role of Cyclosporine in Gingival Hyperplasia: an in Vitro Study on Gingival Fibroblasts
International Journal of Molecular Sciences Article Role of Cyclosporine in Gingival Hyperplasia: An In Vitro Study on Gingival Fibroblasts 1, , 2, 3 3 Dorina Lauritano * y , Annalisa Palmieri y, Alberta Lucchese , Dario Di Stasio , Giulia Moreo 1 and Francesco Carinci 4 1 Department of Medicine and Surgery, Centre of Neuroscience of Milan, University of Milano-Bicocca, 20126 Milan, Italy; [email protected] 2 Department of Experimental, Diagnostic and Specialty Medicine, University of Bologna, via Belmoro 8, 40126 Bologna, Italy; [email protected] 3 Multidisciplinary Department of Medical and Dental Specialties, University of Campania-Luigi Vanvitelli, 80138 Naples, Italy; [email protected] (A.L.); [email protected] (D.D.S.) 4 Department of Morphology, Surgery and Experimental Medicine, University of Ferrara, 44121 Ferrara, Italy; [email protected] * Correspondence: [email protected]; Tel.: +39-335-679-0163 These authors contributed equally to this work. y Received: 25 November 2019; Accepted: 13 January 2020; Published: 16 January 2020 Abstract: Background: Gingival hyperplasia could occur after the administration of cyclosporine A. Up to 90% of the patients submitted to immunosuppressant drugs have been reported to suffer from this side effect. The role of fibroblasts in gingival hyperplasia has been widely discussed by literature, showing contrasting results. In order to demonstrate the effect of cyclosporine A on the extracellular matrix component of fibroblasts, we investigated the gene expression profile of human fibroblasts after cyclosporine A administration. Materials and methods: Primary gingival fibroblasts were stimulated with 1000 ng/mL cyclosporine A solution for 16 h. Gene expression levels of 57 genes belonging to the “Extracellular Matrix and Adhesion Molecules” pathway were analyzed using real-time PCR in treated cells, compared to untreated cells used as control. -

A Single-Cell Transcriptional Atlas Identifies Extensive Heterogeneity in the Cellular Composition of Tendons
bioRxiv preprint doi: https://doi.org/10.1101/801266; this version posted October 10, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. A single-cell transcriptional atlas identifies extensive heterogeneity in the cellular composition of tendons Jacob B Swanson1, Andrea J De Micheli2, Nathaniel P Disser1, Leandro M Martinez1, Nicholas R Walker1,3, Benjamin D Cosgrove2, Christopher L Mendias1,3,* 1Hospital for Special Surgery, New York, NY, USA 2Meining School of Biomedical Engineering, Cornell University, Ithaca, NY, USA 3Department of Physiology and Biophysics, Weill Cornell Medical College, New York, NY, USA *Corresponding Author Christopher Mendias, PhD Hospital for Special Surgery 535 E 70th St New York, NY 10021 USA +1 212-606-1785 [email protected] Keywords: tenocyte; tendon fibroblast; pericyte; single-cell RNA sequencing bioRxiv preprint doi: https://doi.org/10.1101/801266; this version posted October 10, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Tendon is a dense, hypocellular connective tissue that transmits forces between muscles and bones. Cellular heterogeneity is increasingly recognized as an important factor in the biological basis of tissue homeostasis and disease, but little is known about the diversity of cells that populate tendon. Our objective was to explore the heterogeneity of cells in mouse Achilles tendons using single-cell RNA sequencing. We identified 13 unique cell types in tendons, including 4 previously undescribed populations of fibroblasts. -

Linc-DYNC2H1-4 Promotes EMT and CSC Phenotypes by Acting As a Sponge of Mir-145 in Pancreatic Cancer Cells
Citation: Cell Death and Disease (2017) 8, e2924; doi:10.1038/cddis.2017.311 OPEN Official journal of the Cell Death Differentiation Association www.nature.com/cddis Linc-DYNC2H1-4 promotes EMT and CSC phenotypes by acting as a sponge of miR-145 in pancreatic cancer cells Yuran Gao1, Zhicheng Zhang2,3, Kai Li1,3, Liying Gong1, Qingzhu Yang1, Xuemei Huang1, Chengcheng Hong1, Mingfeng Ding*,2 and Huanjie Yang*,1 The acquisition of epithelial–mesenchymal transition (EMT) and/or existence of a sub-population of cancer stem-like cells (CSC) are associated with malignant behavior and chemoresistance. To identify which factor could promote EMT and CSC formation and uncover the mechanistic role of such factor is important for novel and targeted therapies. In the present study, we found that the long intergenic non-coding RNA linc-DYNC2H1-4 was upregulated in pancreatic cancer cell line BxPC-3-Gem with acquired gemcitabine resistance. Knockdown of linc-DYNC2H1-4 decreased the invasive behavior of BxPC-3-Gem cells while ectopic expression of linc-DYNC2H1-4 promoted the acquisition of EMT and stemness of the parental sensitive cells. Linc-DYNC2H1-4 upregulated ZEB1, the EMT key player, which led to upregulation and downregulation of its targets vimentin and E-cadherin respectively, as well as enhanced the expressions of CSC makers Lin28, Nanog, Sox2 and Oct4. Linc-DYNC2H1-4 is mainly located in the cytosol. Mechanically, it could sponge miR-145 that targets ZEB1, Lin28, Nanog, Sox2, Oct4 to restore these EMT and CSC-associated genes expressions. We proved that MMP3, the nearby gene of linc-DYNC2H1-4 in the sense strand, was also a target of miR-145. -

A Collagen-Remodeling Gene Signature Regulated by TGF-B Signaling Is Associated with Metastasis and Poor Survival in Serous Ovarian Cancer
Published OnlineFirst November 11, 2013; DOI: 10.1158/1078-0432.CCR-13-1256 Clinical Cancer Imaging, Diagnosis, Prognosis Research A Collagen-Remodeling Gene Signature Regulated by TGF-b Signaling Is Associated with Metastasis and Poor Survival in Serous Ovarian Cancer Dong-Joo Cheon1, Yunguang Tong2, Myung-Shin Sim7, Judy Dering6, Dror Berel3, Xiaojiang Cui1, Jenny Lester1, Jessica A. Beach1,5, Mourad Tighiouart3, Ann E. Walts4, Beth Y. Karlan1,6, and Sandra Orsulic1,6 Abstract Purpose: To elucidate molecular pathways contributing to metastatic cancer progression and poor clinical outcome in serous ovarian cancer. Experimental Design: Poor survival signatures from three different serous ovarian cancer datasets were compared and a common set of genes was identified. The predictive value of this gene signature was validated in independent datasets. The expression of the signature genes was evaluated in primary, metastatic, and/or recurrent cancers using quantitative PCR and in situ hybridization. Alterations in gene expression by TGF-b1 and functional consequences of loss of COL11A1 were evaluated using pharmacologic and knockdown approaches, respectively. Results: We identified and validated a 10-gene signature (AEBP1, COL11A1, COL5A1, COL6A2, LOX, POSTN, SNAI2, THBS2, TIMP3, and VCAN) that is associated with poor overall survival (OS) in patients with high-grade serous ovarian cancer. The signature genes encode extracellular matrix proteins involved in collagen remodeling. Expression of the signature genes is regulated by TGF-b1 signaling and is enriched in metastases in comparison with primary ovarian tumors. We demonstrate that levels of COL11A1, one of the signature genes, continuously increase during ovarian cancer disease progression, with the highest expression in recurrent metastases. -

Suramin Inhibits Osteoarthritic Cartilage Degradation by Increasing Extracellular Levels
Molecular Pharmacology Fast Forward. Published on August 10, 2017 as DOI: 10.1124/mol.117.109397 This article has not been copyedited and formatted. The final version may differ from this version. MOL #109397 Suramin inhibits osteoarthritic cartilage degradation by increasing extracellular levels of chondroprotective tissue inhibitor of metalloproteinases 3 (TIMP-3). Anastasios Chanalaris, Christine Doherty, Brian D. Marsden, Gabriel Bambridge, Stephen P. Wren, Hideaki Nagase, Linda Troeberg Arthritis Research UK Centre for Osteoarthritis Pathogenesis, Kennedy Institute of Downloaded from Rheumatology, University of Oxford, Roosevelt Drive, Headington, Oxford OX3 7FY, UK (A.C., C.D., G.B., H.N., L.T.); Alzheimer’s Research UK Oxford Drug Discovery Institute, University of Oxford, Oxford, OX3 7FZ, UK (S.P.W.); Structural Genomics Consortium, molpharm.aspetjournals.org University of Oxford, Old Road Campus Research Building, Old Road Campus, Roosevelt Drive, Headington, Oxford, OX3 7DQ (BDM). at ASPET Journals on September 29, 2021 1 Molecular Pharmacology Fast Forward. Published on August 10, 2017 as DOI: 10.1124/mol.117.109397 This article has not been copyedited and formatted. The final version may differ from this version. MOL #109397 Running title: Repurposing suramin to inhibit osteoarthritic cartilage loss. Corresponding author: Linda Troeberg Address: Kennedy Institute of Rheumatology, University of Oxford, Roosevelt Drive, Headington, Oxford OX3 7FY, UK Phone number: +44 (0)1865 612600 E-mail: [email protected] Downloaded -

Gene Symbol Category ACAN ECM ADAM10 ECM Remodeling-Related ADAM11 ECM Remodeling-Related ADAM12 ECM Remodeling-Related ADAM15 E
Supplementary Material (ESI) for Integrative Biology This journal is (c) The Royal Society of Chemistry 2010 Gene symbol Category ACAN ECM ADAM10 ECM remodeling-related ADAM11 ECM remodeling-related ADAM12 ECM remodeling-related ADAM15 ECM remodeling-related ADAM17 ECM remodeling-related ADAM18 ECM remodeling-related ADAM19 ECM remodeling-related ADAM2 ECM remodeling-related ADAM20 ECM remodeling-related ADAM21 ECM remodeling-related ADAM22 ECM remodeling-related ADAM23 ECM remodeling-related ADAM28 ECM remodeling-related ADAM29 ECM remodeling-related ADAM3 ECM remodeling-related ADAM30 ECM remodeling-related ADAM5 ECM remodeling-related ADAM7 ECM remodeling-related ADAM8 ECM remodeling-related ADAM9 ECM remodeling-related ADAMTS1 ECM remodeling-related ADAMTS10 ECM remodeling-related ADAMTS12 ECM remodeling-related ADAMTS13 ECM remodeling-related ADAMTS14 ECM remodeling-related ADAMTS15 ECM remodeling-related ADAMTS16 ECM remodeling-related ADAMTS17 ECM remodeling-related ADAMTS18 ECM remodeling-related ADAMTS19 ECM remodeling-related ADAMTS2 ECM remodeling-related ADAMTS20 ECM remodeling-related ADAMTS3 ECM remodeling-related ADAMTS4 ECM remodeling-related ADAMTS5 ECM remodeling-related ADAMTS6 ECM remodeling-related ADAMTS7 ECM remodeling-related ADAMTS8 ECM remodeling-related ADAMTS9 ECM remodeling-related ADAMTSL1 ECM remodeling-related ADAMTSL2 ECM remodeling-related ADAMTSL3 ECM remodeling-related ADAMTSL4 ECM remodeling-related ADAMTSL5 ECM remodeling-related AGRIN ECM ALCAM Cell-cell adhesion ANGPT1 Soluble factors and receptors -

A Meta-Analysis of the Inhibin Network Reveals Prognostic Value in Multiple Solid Tumors
bioRxiv preprint doi: https://doi.org/10.1101/2020.06.25.171942; this version posted June 27, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 A meta-analysis of the inhibin network reveals prognostic value in multiple solid tumors Eduardo Listik1†, Ben Horst1,2†, Alex Seok Choi1, Nam. Y. Lee3, Balázs Győrffy4 and Karthikeyan Mythreye1 Affiliations: 1Department of Pathology, University of Alabama at Birmingham, Birmingham AL, USA, 35294. 2Department of Chemistry and Biochemistry, University of South Carolina, Columbia SC, USA, 29208. 3 Division of Pharmacology, Chemistry and Biochemistry, College of Medicine, University of Arizona, Tucson, AZ, 85721, USA. 4 TTK Cancer Biomarker Research Group, Institute of Enzymology, and Semmelweis University Department of Bioinformatics and 2nd Department of Pediatrics, Budapest, Hungary. Corresponding author: Karthikeyan Mythreye, Ph.D. Division of Molecular and Cellular Pathology, Department of Pathology, The University of Alabama at Birmingham. WTI 320B, 1824 Sixth Avenue South, Birmingham, AL, USA, 35294. Phone : +1 205.934.2746 E-mail : [email protected] †The authors contributed equally to this work. bioRxiv preprint doi: https://doi.org/10.1101/2020.06.25.171942; this version posted June 27, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. -

Investigation of COVID-19 Comorbidities Reveals Genes and Pathways Coincident with the SARS-Cov-2 Viral Disease
bioRxiv preprint doi: https://doi.org/10.1101/2020.09.21.306720; this version posted September 21, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Title: Investigation of COVID-19 comorbidities reveals genes and pathways coincident with the SARS-CoV-2 viral disease. Authors: Mary E. Dolan1*,2, David P. Hill1,2, Gaurab Mukherjee2, Monica S. McAndrews2, Elissa J. Chesler2, Judith A. Blake2 1 These authors contributed equally and should be considered co-first authors * Corresponding author [email protected] 2 The Jackson Laboratory, 600 Main St, Bar Harbor, ME 04609, USA Abstract: The emergence of the SARS-CoV-2 virus and subsequent COVID-19 pandemic initiated intense research into the mechanisms of action for this virus. It was quickly noted that COVID-19 presents more seriously in conjunction with other human disease conditions such as hypertension, diabetes, and lung diseases. We conducted a bioinformatics analysis of COVID-19 comorbidity-associated gene sets, identifying genes and pathways shared among the comorbidities, and evaluated current knowledge about these genes and pathways as related to current information about SARS-CoV-2 infection. We performed our analysis using GeneWeaver (GW), Reactome, and several biomedical ontologies to represent and compare common COVID- 19 comorbidities. Phenotypic analysis of shared genes revealed significant enrichment for immune system phenotypes and for cardiovascular-related phenotypes, which might point to alleles and phenotypes in mouse models that could be evaluated for clues to COVID-19 severity. -

Effect of Nanoparticles on the Expression and Activity of Matrix Metalloproteinases
Nanotechnol Rev 2018; 7(6): 541–553 Review Magdalena Matysiak-Kucharek*, Magdalena Czajka, Krzysztof Sawicki, Marcin Kruszewski and Lucyna Kapka-Skrzypczak Effect of nanoparticles on the expression and activity of matrix metalloproteinases https://doi.org/10.1515/ntrev-2018-0110 Received September 14, 2018; accepted October 11, 2018; previously 1 Introduction published online November 15, 2018 Matrix metallopeptidases, commonly known as matrix Abstract: Matrix metallopeptidases, commonly known metalloproteinases (MMPs), are zinc-dependent proteo- as matrix metalloproteinases (MMPs), are a group of pro- lytic enzymes whose primary function is the degradation teolytic enzymes whose main function is the remodeling and remodeling of extracellular matrix (ECM) compo- of the extracellular matrix. Changes in the activity of nents. ECM is a complex, dynamic structure that condi- these enzymes are observed in many pathological states, tions the proper tissue architecture. MMPs by digesting including cancer metastases. An increasing body of evi- ECM proteins eliminate structural barriers and allow dence indicates that nanoparticles (NPs) can lead to the cell migration. Moreover, by hydrolyzing extracellularly deregulation of MMP expression and/or activity both in released proteins, MMPs can change the activity of many vitro and in vivo. In this work, we summarized the current signal peptides, such as growth factors, cytokines, and state of knowledge on the impact of NPs on MMPs. The chemokines. MMPs are involved in many physiological literature analysis showed that the impact of NPs on MMP processes, such as embryogenesis, reproduction cycle, or expression and/or activity is inconclusive. NPs exhibit wound healing; however, their increased activity is also both stimulating and inhibitory effects, which might be associated with a number of pathological conditions, such dependent on multiple factors, such as NP size and coat- as diabetes, cardiovascular diseases and neurodegenera- ing or a cellular model used in the research. -

Polymorphisms of the Matrix Metalloproteinase Genes
www.nature.com/scientificreports OPEN Polymorphisms of the matrix metalloproteinase genes are associated with essential hypertension in a Caucasian population of Central Russia Maria Moskalenko1, Irina Ponomarenko1, Evgeny Reshetnikov1*, Volodymyr Dvornyk2 & Mikhail Churnosov1 This study aimed to determine possible association of eight polymorphisms of seven MMP genes with essential hypertension (EH) in a Caucasian population of Central Russia. Eight SNPs of the MMP1, MMP2, MMP3, MMP7, MMP8, MMP9, and MMP12 genes and their gene–gene (epistatic) interactions were analyzed for association with EH in a cohort of 939 patients and 466 controls using logistic regression and assuming additive, recessive, and dominant genetic models. The functional signifcance of the polymorphisms associated with EH and 114 variants linked to them (r2 ≥ 0.8) was analyzed in silico. Allele G of rs11568818 MMP7 was associated with EH according to all three genetic models (OR = 0.58–0.70, pperm = 0.01–0.03). The above eight SNPs were associated with the disorder within 12 most signifcant epistatic models (OR = 1.49–1.93, pperm < 0.02). Loci rs1320632 MMP8 and rs11568818 MMP7 contributed to the largest number of the models (12 and 10, respectively). The EH-associated loci and 114 SNPs linked to them had non-synonymous, regulatory, and eQTL signifcance for 15 genes, which contributed to the pathways related to metalloendopeptidase activity, collagen degradation, and extracellular matrix disassembly. In summary, eight studied SNPs of MMPs genes were associated with EH in the Caucasian population of Central Russia. Cardiovascular diseases are a global problem of modern healthcare and the second most common cause of total mortality1,2. -
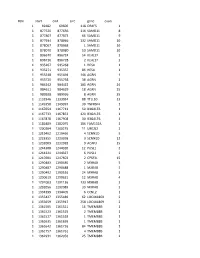
Chr Start End Size Gene Exon 1 69482 69600 118 OR4F5 1 1 877520
#chr start end size gene exon 1 69482 69600 118 OR4F5 1 1 877520 877636 116 SAMD11 8 1 877807 877873 66 SAMD11 9 1 877934 878066 132 SAMD11 10 1 878067 878068 1 SAMD11 10 1 878070 878080 10 SAMD11 10 1 896670 896724 54 KLHL17 2 1 896726 896728 2 KLHL17 2 1 935267 935268 1 HES4 1 1 935271 935357 86 HES4 1 1 955548 955694 146 AGRN 1 1 955720 955758 38 AGRN 1 1 984242 984422 180 AGRN 24 1 984611 984629 18 AGRN 25 1 989928 989936 8 AGRN 35 1 1132946 1133034 88 TTLL10 13 1 1149358 1149397 39 TNFRSF4 1 1 1167654 1167713 59 B3GALT6 1 1 1167733 1167853 120 B3GALT6 1 1 1167878 1167908 30 B3GALT6 1 1 1181889 1182075 186 FAM132A 1 1 1200204 1200215 11 UBE2J2 2 1 1219462 1219466 4 SCNN1D 5 1 1223355 1223358 3 SCNN1D 12 1 1232009 1232018 9 ACAP3 15 1 1244308 1244320 12 PUSL1 2 1 1244321 1244327 6 PUSL1 2 1 1247601 1247603 2 CPSF3L 15 1 1290483 1290485 2 MXRA8 5 1 1290487 1290488 1 MXRA8 5 1 1290492 1290516 24 MXRA8 5 1 1290619 1290631 12 MXRA8 4 1 1291003 1291136 133 MXRA8 3 1 1292056 1292089 33 MXRA8 2 1 1334399 1334405 6 CCNL2 1 1 1355427 1355489 62 LOC441869 2 1 1355659 1355917 258 LOC441869 2 1 1361505 1361521 16 TMEM88B 1 1 1361523 1361525 2 TMEM88B 1 1 1361527 1361528 1 TMEM88B 1 1 1361635 1361636 1 TMEM88B 1 1 1361642 1361726 84 TMEM88B 1 1 1361757 1361761 4 TMEM88B 1 1 1362931 1362956 25 TMEM88B 2 1 1374780 1374838 58 VWA1 3 1 1374841 1374842 1 VWA1 3 1 1374934 1375100 166 VWA1 3 1 1389738 1389747 9 ATAD3C 4 1 1389751 1389816 65 ATAD3C 4 1 1389830 1389849 19 ATAD3C 4 1 1390835 1390837 2 ATAD3C 5 1 1407260 1407331 71 ATAD3B 1 1 1407340 1407474 -

FOP) Can Be Rescued by the Drug Candidate Saracatinib
Stem Cell Reviews and Reports https://doi.org/10.1007/s12015-020-10103-9 ActivinA Induced SMAD1/5 Signaling in an iPSC Derived EC Model of Fibrodysplasia Ossificans Progressiva (FOP) Can Be Rescued by the Drug Candidate Saracatinib Susanne Hildebrandt1,2,3 & Branka Kampfrath1 & Kristin Fischer3,4 & Laura Hildebrand2,3 & Julia Haupt1 & Harald Stachelscheid3,4 & Petra Knaus 1,2 Accepted: 1 December 2020 # The Author(s) 2021 Abstract Balanced signal transduction is crucial in tissue patterning, particularly in the vasculature. Heterotopic ossification (HO) is tightly linked to vascularization with increased vessel number in hereditary forms of HO, such as Fibrodysplasia ossificans progressiva (FOP). FOP is caused by mutations in the BMP type I receptor ACVR1 leading to aberrant SMAD1/5 signaling in response to ActivinA. Whether observed vascular phenotype in human FOP lesions is connected to aberrant ActivinA signaling is unknown. Blocking of ActivinA prevents HO in FOP mice indicating a central role of the ligand in FOP. Here, we established a new FOP endothelial cell model generated from induced pluripotent stem cells (iECs) to study ActivinA signaling. FOP iECs recapitulate pathogenic ActivinA/SMAD1/5 signaling. Whole transcriptome analysis identified ActivinA mediated activation of the BMP/ NOTCH pathway exclusively in FOP iECs, which was rescued to WT transcriptional levels by the drug candidate Saracatinib. We propose that ActivinA causes transcriptional pre-patterning of the FOP endothelium, which might contribute to differential vascularity in FOP lesions compared to non-hereditary HO. Keywords FOP . BMP-receptor . Activin . iPSCs . Human endothelial cells . HO . Saracatinib Introduction pathological conditions such as cancer and chronic inflamma- tion [2].