KO Kidney.Xlsx
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

IMPDH2: a New Gene Associated with Dominant Juvenile-Onset Dystonia-Tremor Disorder
www.nature.com/ejhg BRIEF COMMUNICATION OPEN IMPDH2: a new gene associated with dominant juvenile-onset dystonia-tremor disorder 1,8 1,8 2 3 1,4 2 5 Anna Kuukasjärvi , Juan✉ C. Landoni , Jyrki Kaukonen , Mika Juhakoski , Mari Auranen , Tommi Torkkeli , Vidya Velagapudi and Anu Suomalainen 1,6,7 © The Author(s) 2021 The aetiology of dystonia disorders is complex, and next-generation sequencing has become a useful tool in elucidating the variable genetic background of these diseases. Here we report a deleterious heterozygous truncating variant in the inosine monophosphate dehydrogenasegene(IMPDH2) by whole-exome sequencing, co-segregating with a dominantly inherited dystonia-tremor disease in a large Finnish family. We show that the defect results in degradation of the gene product, causing IMPDH2 deficiency in patient cells. IMPDH2 is the first and rate-limiting enzyme in the de novo biosynthesis of guanine nucleotides, a dopamine synthetic pathway previously linked to childhood or adolescence-onset dystonia disorders. We report IMPDH2 as a new gene to the dystonia disease entity. The evidence underlines the important link between guanine metabolism, dopamine biosynthesis and dystonia. European Journal of Human Genetics; https://doi.org/10.1038/s41431-021-00939-1 INTRODUCTION The disease-onset was between 9 and 20 years of age. Table 1 Dystonias are rare movement disorders characterised by sustained or summarises the clinical presentations. intermittent muscle contractions causing abnormal, often repetitive, movements and/or postures. Dystonia can manifest as an isolated Case report symptom or combined with e.g. parkinsonism or myoclonus [1]. While Patient II-6 is a 46-year-old woman. -

Chuanxiong Rhizoma Compound on HIF-VEGF Pathway and Cerebral Ischemia-Reperfusion Injury’S Biological Network Based on Systematic Pharmacology
ORIGINAL RESEARCH published: 25 June 2021 doi: 10.3389/fphar.2021.601846 Exploring the Regulatory Mechanism of Hedysarum Multijugum Maxim.-Chuanxiong Rhizoma Compound on HIF-VEGF Pathway and Cerebral Ischemia-Reperfusion Injury’s Biological Network Based on Systematic Pharmacology Kailin Yang 1†, Liuting Zeng 1†, Anqi Ge 2†, Yi Chen 1†, Shanshan Wang 1†, Xiaofei Zhu 1,3† and Jinwen Ge 1,4* Edited by: 1 Takashi Sato, Key Laboratory of Hunan Province for Integrated Traditional Chinese and Western Medicine on Prevention and Treatment of 2 Tokyo University of Pharmacy and Life Cardio-Cerebral Diseases, Hunan University of Chinese Medicine, Changsha, China, Galactophore Department, The First 3 Sciences, Japan Hospital of Hunan University of Chinese Medicine, Changsha, China, School of Graduate, Central South University, Changsha, China, 4Shaoyang University, Shaoyang, China Reviewed by: Hui Zhao, Capital Medical University, China Background: Clinical research found that Hedysarum Multijugum Maxim.-Chuanxiong Maria Luisa Del Moral, fi University of Jaén, Spain Rhizoma Compound (HCC) has de nite curative effect on cerebral ischemic diseases, *Correspondence: such as ischemic stroke and cerebral ischemia-reperfusion injury (CIR). However, its Jinwen Ge mechanism for treating cerebral ischemia is still not fully explained. [email protected] †These authors share first authorship Methods: The traditional Chinese medicine related database were utilized to obtain the components of HCC. The Pharmmapper were used to predict HCC’s potential targets. Specialty section: The CIR genes were obtained from Genecards and OMIM and the protein-protein This article was submitted to interaction (PPI) data of HCC’s targets and IS genes were obtained from String Ethnopharmacology, a section of the journal database. -

Type of the Paper (Article
Supplementary Material A Proteomics Study on the Mechanism of Nutmeg-induced Hepatotoxicity Wei Xia 1, †, Zhipeng Cao 1, †, Xiaoyu Zhang 1 and Lina Gao 1,* 1 School of Forensic Medicine, China Medical University, Shenyang 110122, P. R. China; lessen- [email protected] (W.X.); [email protected] (Z.C.); [email protected] (X.Z.) † The authors contributed equally to this work. * Correspondence: [email protected] Figure S1. Table S1. Peptide fraction separation liquid chromatography elution gradient table. Time (min) Flow rate (mL/min) Mobile phase A (%) Mobile phase B (%) 0 1 97 3 10 1 95 5 30 1 80 20 48 1 60 40 50 1 50 50 53 1 30 70 54 1 0 100 1 Table 2. Liquid chromatography elution gradient table. Time (min) Flow rate (nL/min) Mobile phase A (%) Mobile phase B (%) 0 600 94 6 2 600 83 17 82 600 60 40 84 600 50 50 85 600 45 55 90 600 0 100 Table S3. The analysis parameter of Proteome Discoverer 2.2. Item Value Type of Quantification Reporter Quantification (TMT) Enzyme Trypsin Max.Missed Cleavage Sites 2 Precursor Mass Tolerance 10 ppm Fragment Mass Tolerance 0.02 Da Dynamic Modification Oxidation/+15.995 Da (M) and TMT /+229.163 Da (K,Y) N-Terminal Modification Acetyl/+42.011 Da (N-Terminal) and TMT /+229.163 Da (N-Terminal) Static Modification Carbamidomethyl/+57.021 Da (C) 2 Table S4. The DEPs between the low-dose group and the control group. Protein Gene Fold Change P value Trend mRNA H2-K1 0.380 0.010 down Glutamine synthetase 0.426 0.022 down Annexin Anxa6 0.447 0.032 down mRNA H2-D1 0.467 0.002 down Ribokinase Rbks 0.487 0.000 -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -
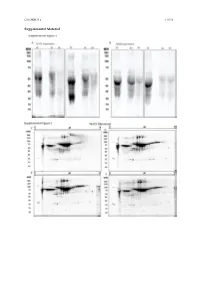
Type of the Paper (Article
Cells 2020, 9, x 1 of 19 Supplemental Material Cells 2020, 9, x 2 of 19 Figure S1. Secretome enrichment: protocol optimization. 1D SDS-PAGE documentation of washing steps: Culture medium was substituted with FCS-free medium, which was changed every 2 h. The supernatants were then collected, and the proteins isolated and separated in 1D SDS-PAGE ((A) TK173 and (B) TK188). Proteins were stained with Flamingo fluorescent gel stain. Two-dimensional pattern of the proteins isolated from supernatant of TK173, (C) 2 h, (D) 4 h, (E) 6 h, and (F) 8 h after changing to FCS-free medium. (G) Cell secretome collected 24 h after elimination of the contaminating FCS-proteins with different washing steps. Proteins were stained with Flamingo fluorescent gel stain. Cells 2020, 9, x 3 of 19 Figure S2. 2-DE reference maps of secretomes; 150 μg proteins were loaded on an 11 cm IPG strip with a linear pH gradient PI 5–8 for IEF; 12% SDS-polyacrylamide gels were used for the second dimension. Proteins were stained with Flamingo fluorescent gel stain. Identified spots were assigned a number corresponding to that in their table. 2-DE maps from secretome of (A) TK173 control and (B) TGFβ1- treated ones. The 2-DE patterns revealed an alteration of secretome in stimulated TK173. Secretome patterns from TK173 treated with (C) ANG II and (D) PDGF. Cells 2020, 9, x 4 of 19 A Figure S3. Classification of the differentially expressed proteins upon ANG II, TGFβ1, or PDGF treatment in TK173. (A) Bar charts of the cellular component analyzed by STRAP biological function analysis in which the identified proteins from all treatments in both cell types are involved. -

Supplementary Methods
Supplementary methods Human lung tissues and tissue microarray (TMA) All human tissues were obtained from the Lung Cancer Specialized Program of Research Excellence (SPORE) Tissue Bank at the M.D. Anderson Cancer Center (Houston, TX). A collection of 26 lung adenocarcinomas and 24 non-tumoral paired tissues were snap-frozen and preserved in liquid nitrogen for total RNA extraction. For each tissue sample, the percentage of malignant tissue was calculated and the cellular composition of specimens was determined by histological examination (I.I.W.) following Hematoxylin-Eosin (H&E) staining. All malignant samples retained contained more than 50% tumor cells. Specimens resected from NSCLC stages I-IV patients who had no prior chemotherapy or radiotherapy were used for TMA analysis by immunohistochemistry. Patients who had smoked at least 100 cigarettes in their lifetime were defined as smokers. Samples were fixed in formalin, embedded in paraffin, stained with H&E, and reviewed by an experienced pathologist (I.I.W.). The 413 tissue specimens collected from 283 patients included 62 normal bronchial epithelia, 61 bronchial hyperplasias (Hyp), 15 squamous metaplasias (SqM), 9 squamous dysplasias (Dys), 26 carcinomas in situ (CIS), as well as 98 squamous cell carcinomas (SCC) and 141 adenocarcinomas. Normal bronchial epithelia, hyperplasia, squamous metaplasia, dysplasia, CIS, and SCC were considered to represent different steps in the development of SCCs. All tumors and lesions were classified according to the World Health Organization (WHO) 2004 criteria. The TMAs were prepared with a manual tissue arrayer (Advanced Tissue Arrayer ATA100, Chemicon International, Temecula, CA) using 1-mm-diameter cores in triplicate for tumors and 1.5 to 2-mm cores for normal epithelial and premalignant lesions. -

(10) Patent No.: US 8119385 B2
US008119385B2 (12) United States Patent (10) Patent No.: US 8,119,385 B2 Mathur et al. (45) Date of Patent: Feb. 21, 2012 (54) NUCLEICACIDS AND PROTEINS AND (52) U.S. Cl. ........................................ 435/212:530/350 METHODS FOR MAKING AND USING THEMI (58) Field of Classification Search ........................ None (75) Inventors: Eric J. Mathur, San Diego, CA (US); See application file for complete search history. Cathy Chang, San Diego, CA (US) (56) References Cited (73) Assignee: BP Corporation North America Inc., Houston, TX (US) OTHER PUBLICATIONS c Mount, Bioinformatics, Cold Spring Harbor Press, Cold Spring Har (*) Notice: Subject to any disclaimer, the term of this bor New York, 2001, pp. 382-393.* patent is extended or adjusted under 35 Spencer et al., “Whole-Genome Sequence Variation among Multiple U.S.C. 154(b) by 689 days. Isolates of Pseudomonas aeruginosa” J. Bacteriol. (2003) 185: 1316 1325. (21) Appl. No.: 11/817,403 Database Sequence GenBank Accession No. BZ569932 Dec. 17. 1-1. 2002. (22) PCT Fled: Mar. 3, 2006 Omiecinski et al., “Epoxide Hydrolase-Polymorphism and role in (86). PCT No.: PCT/US2OO6/OOT642 toxicology” Toxicol. Lett. (2000) 1.12: 365-370. S371 (c)(1), * cited by examiner (2), (4) Date: May 7, 2008 Primary Examiner — James Martinell (87) PCT Pub. No.: WO2006/096527 (74) Attorney, Agent, or Firm — Kalim S. Fuzail PCT Pub. Date: Sep. 14, 2006 (57) ABSTRACT (65) Prior Publication Data The invention provides polypeptides, including enzymes, structural proteins and binding proteins, polynucleotides US 201O/OO11456A1 Jan. 14, 2010 encoding these polypeptides, and methods of making and using these polynucleotides and polypeptides. -

Cell-Specific Proteomic Analysis in Caenorhabditis Elegans
Supporting Information Appendix (265 Pages) for Cell-Specific Proteomic Analysis in Caenorhabditis elegans Authors: Kai P. Yueta, Meenakshi K. Domab, c, John T. Ngoa, 2, Michael J. Sweredoskid, Robert L. J. Grahamd, 3, Annie Moradiand, Sonja Hessd, Erin M. Schumane, Paul W. Sternbergb,c and David A. Tirrella,1 Author Affiliations: aDivision of Chemistry and Chemical Engineering, California Institute of Technology, Pasadena, California, United States of America bDivision of Biology and Biological Engineering, California Institute of Technology, Pasadena, California, United States of America cHoward Hughes Medical Institute, California Institute of Technology, Pasadena, California, United States of America dProteome Exploration Laboratory, Beckman Institute, California Institute of Technology, Pasadena, California, United States of America eMax Planck Institute for Brain Research, Frankfurt am Main, Germany 1To whom correspondence may be addressed. 2Current Address: Department of Pharmacology, University of California, San Diego, La Jolla, California, United States of America 3Current Address: Faculty of Medical and Human Sciences, University of Manchester, Manchester, United Kingdom Supporting Information Materials and Methods - Adenosine Triphosphate (ATP)-Pyrophosphate (PPi) Exchange Assay - Chloroform/Methanol Precipitation - Enrichment of p-Azido-L-Phenylalanine-Labeled Proteins - Fluorescence Microscopy of Live C. elegans - Fluorescence Microscopy of p-Azido-L-Phenylalanine-Labeled Proteins in Fixed C. elegans - In-Gel Fluorescence -

Supplementary Data.Xlsx
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is © The Royal Society of Chemistry 2016 Average Average spectral spectral Fold UniProt IDGene Protein Name counts- counts- enrichm negative positive ent sample sample P12821 ACE HUMAN - ACE Angiotensin-converting enzyme 0 79.75 #DIV/0! Q71U36 TBA1A HUMAN - TUBA1A Tubulin alpha-1A chain 0 59.5 #DIV/0! P17812 PYRG1 HUMAN - CTPS1 CTP synthase 1 0 43.5 #DIV/0! P23921 RIR1 HUMAN - RRM1 Ribonucleoside-diphosphate reductase large subunit 0 35 #DIV/0! P49915GUAA HUMAN - GMPS GMP synthase 0 30.5 #DIV/0! P30153 2AAA HUMAN - PPP2R1A Serine/threonine-protein phosphatase 2A 65 kDa0 regulatory subunit29 A#DIV/0! alpha isoform P55786 PSA HUMAN - NPEPPS Puromycin-sensitive aminopeptidase 0 28.75 #DIV/0! O43143 DHX15 HUMAN - DHX15 Putative pre-mRNA-splicing factor ATP-dependent RNA0 helicase28.25 DHX15#DIV/0! P15170 ERF3A HUMAN - GSPT1 Eukaryotic peptide chain release factor GTP-binding0 subunit ERF3A24.75 #DIV/0! P09874PARP1HUMAN - PARP1 Poly 0 23.5 #DIV/0! Q9BXJ9 NAA15 HUMAN - NAA15 N-alpha-acetyltransferase 15, NatA auxiliary subunit0 23 #DIV/0! B0V043 B0V043 HUMAN - VARS Valyl-tRNA synthetase 0 20 #DIV/0! Q86VP6 CAND1 HUMAN - CAND1 Cullin-associated NEDD8-dissociated protein 1 0 19.5 #DIV/0! P04080CYTB HUMAN - CSTB Cystatin-B 0 19 #DIV/0! Q93009 UBP7 HUMAN - USP7 Ubiquitin carboxyl-terminal hydrolase 7 0 18 #DIV/0! Q9Y2L1 RRP44 HUMAN - DIS3 Exosome complex exonuclease RRP44 0 18 #DIV/0! Q13748 TBA3C HUMAN - TUBA3D Tubulin alpha-3C/D chain 0 18 #DIV/0! P29144 TPP2 HUMAN -

Inda, María Carolina. 2017 03 15
Tesis Doctoral Caracterización de distintas fuentes de cAMP en la señalización del GPCR CRHR1 Inda, María Carolina 2017-03-15 Este documento forma parte de la colección de tesis doctorales y de maestría de la Biblioteca Central Dr. Luis Federico Leloir, disponible en digital.bl.fcen.uba.ar. Su utilización debe ser acompañada por la cita bibliográfica con reconocimiento de la fuente. This document is part of the doctoral theses collection of the Central Library Dr. Luis Federico Leloir, available in digital.bl.fcen.uba.ar. It should be used accompanied by the corresponding citation acknowledging the source. Cita tipo APA: Inda, María Carolina. (2017-03-15). Caracterización de distintas fuentes de cAMP en la señalización del GPCR CRHR1. Facultad de Ciencias Exactas y Naturales. Universidad de Buenos Aires. Cita tipo Chicago: Inda, María Carolina. "Caracterización de distintas fuentes de cAMP en la señalización del GPCR CRHR1". Facultad de Ciencias Exactas y Naturales. Universidad de Buenos Aires. 2017-03- 15. Dirección: Biblioteca Central Dr. Luis F. Leloir, Facultad de Ciencias Exactas y Naturales, Universidad de Buenos Aires. Contacto: [email protected] Intendente Güiraldes 2160 - C1428EGA - Tel. (++54 +11) 4789-9293 UNIVERSIDAD DE BUENOS AIRES Facultad de Ciencias Exactas y Naturales Caracterización de distintas fuentes de cAMP en la señalización del GPCR CRHR1 Tesis presentada para optar al título de Doctor de la Universidad de Buenos Aires en el área de Ciencias Biológicas Lic. María Carolina Inda Directora de tesis: Dr. Susana Silberstein Cuña Consejero de Estudios: Dr. Eduardo Arzt Instituto de Investigación en Biomedicina de Buenos Aires CONICET - Partner Institute of the Max Planck Society Buenos Aires, 2017 Gracias Susana por la confianza y el apoyo para trabajar todos estos años. -

Deciphering the Ovarian Proteomic Impacts of Obesity
Iowa State University Capstones, Theses and Graduate Theses and Dissertations Dissertations 2019 Deciphering the ovarian proteomic impacts of obesity Kendra Leah Clark Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/etd Part of the Genetics Commons Recommended Citation Clark, Kendra Leah, "Deciphering the ovarian proteomic impacts of obesity" (2019). Graduate Theses and Dissertations. 17659. https://lib.dr.iastate.edu/etd/17659 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. Deciphering the ovarian proteomic impacts of obesity by Kendra Leah Clark A dissertation submitted to the graduate faculty in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Major: Genetics and Genomics Program of Study Committee: Aileen F. Keating, Major Professor Jeffrey J. Essner Donald S. Sakaguchi Jason W. Ross Geetu Tuteja The student author, whose presentation of the scholarship herein was approved by the program of study committee, is solely responsible for the content of this dissertation. The Graduate College will ensure this dissertation is globally accessible and will not permit alterations after a degree is conferred. Iowa State University Ames, Iowa 2019 Copyright © Kendra Leah Clark, 2019. All rights reserved. ii DEDICATION I dedicate this thesis to my children; whose presence gave me reason. Additionally, I dedicate this work to all the people out there with questionable pasts that everyone gave up on but who never gave up on themselves.