1 T CELL ACTIVATION TRIGGERS REVERSIBLE INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE ASSEMBLY Krisna C. Duong-Ly 1, Yin-Ming Kuo 2
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

KO Kidney.Xlsx
Supplemental Table 18: Dietary Impact on the CGL KO Kidney Sulfhydrome DR/AL Accession Molecular Cysteine Spectral Protein Name Number Alternate ID Weight Residues Count Ratio P‐value Ig gamma‐2A chain C region, A allele P01863 (+1) Ighg 36 kDa 10 C 5.952 0.03767 Heterogeneous nuclear ribonucleoprotein M Q9D0E1 (+1) Hnrnpm 78 kDa 6 C 5.000 0.00595 Phospholipase D3 O35405 Pld3 54 kDa 8 C 4.167 0.04761 Ig kappa chain V‐V region L7 (Fragment) P01642 Gm10881 13 kDa 2 C 2.857 0.01232 UPF0160 protein MYG1, mitochondrial Q9JK81 Myg1 43 kDa 7 C 2.333 0.01613 Copper homeostasis protein cutC homolog Q9D8X1 Cutc 29 kDa 7 C 10.333 0.16419 Corticosteroid‐binding globulin Q06770 Serpina6 45 kDa 3 C 10.333 0.16419 28S ribosomal protein S22, mitochondrial Q9CXW2 Mrps22 41 kDa 2 C 7.333 0.3739 Isoform 3 of Agrin A2ASQ1‐3 Agrn 198 kDa 2 C 7.333 0.3739 3‐oxoacyl‐[acyl‐carrier‐protein] synthase, mitochondrial Q9D404 Oxsm 49 kDa 11 C 7.333 0.3739 Cordon‐bleu protein‐like 1 Q3UMF0 (+3)Cobll1 137 kDa 10 C 5.833 0.10658 ADP‐sugar pyrophosphatase Q9JKX6 Nudt5 24 kDa 5 C 4.167 0.15819 Complement C4‐B P01029 C4b 193 kDa 29 C 3.381 0.23959 Protein‐glutamine gamma‐glutamyltransferase 2 P21981 Tgm2 77 kDa 20 C 3.381 0.23959 Isochorismatase domain‐containing protein 1 Q91V64 Isoc1 32 kDa 5 C 3.333 0.10588 Serpin B8 O08800 Serpinb8 42 kDa 11 C 2.903 0.06902 Heterogeneous nuclear ribonucleoprotein A0 Q9CX86 Hnrnpa0 31 kDa 3 C 2.667 0.5461 Proteasome subunit beta type‐8 P28063 Psmb8 30 kDa 5 C 2.583 0.36848 Ig kappa chain V‐V region MOPC 149 P01636 12 kDa 2 C 2.583 0.36848 -

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

IMPDH2: a New Gene Associated with Dominant Juvenile-Onset Dystonia-Tremor Disorder
www.nature.com/ejhg BRIEF COMMUNICATION OPEN IMPDH2: a new gene associated with dominant juvenile-onset dystonia-tremor disorder 1,8 1,8 2 3 1,4 2 5 Anna Kuukasjärvi , Juan✉ C. Landoni , Jyrki Kaukonen , Mika Juhakoski , Mari Auranen , Tommi Torkkeli , Vidya Velagapudi and Anu Suomalainen 1,6,7 © The Author(s) 2021 The aetiology of dystonia disorders is complex, and next-generation sequencing has become a useful tool in elucidating the variable genetic background of these diseases. Here we report a deleterious heterozygous truncating variant in the inosine monophosphate dehydrogenasegene(IMPDH2) by whole-exome sequencing, co-segregating with a dominantly inherited dystonia-tremor disease in a large Finnish family. We show that the defect results in degradation of the gene product, causing IMPDH2 deficiency in patient cells. IMPDH2 is the first and rate-limiting enzyme in the de novo biosynthesis of guanine nucleotides, a dopamine synthetic pathway previously linked to childhood or adolescence-onset dystonia disorders. We report IMPDH2 as a new gene to the dystonia disease entity. The evidence underlines the important link between guanine metabolism, dopamine biosynthesis and dystonia. European Journal of Human Genetics; https://doi.org/10.1038/s41431-021-00939-1 INTRODUCTION The disease-onset was between 9 and 20 years of age. Table 1 Dystonias are rare movement disorders characterised by sustained or summarises the clinical presentations. intermittent muscle contractions causing abnormal, often repetitive, movements and/or postures. Dystonia can manifest as an isolated Case report symptom or combined with e.g. parkinsonism or myoclonus [1]. While Patient II-6 is a 46-year-old woman. -

35 Disorders of Purine and Pyrimidine Metabolism
35 Disorders of Purine and Pyrimidine Metabolism Georges van den Berghe, M.- Françoise Vincent, Sandrine Marie 35.1 Inborn Errors of Purine Metabolism – 435 35.1.1 Phosphoribosyl Pyrophosphate Synthetase Superactivity – 435 35.1.2 Adenylosuccinase Deficiency – 436 35.1.3 AICA-Ribosiduria – 437 35.1.4 Muscle AMP Deaminase Deficiency – 437 35.1.5 Adenosine Deaminase Deficiency – 438 35.1.6 Adenosine Deaminase Superactivity – 439 35.1.7 Purine Nucleoside Phosphorylase Deficiency – 440 35.1.8 Xanthine Oxidase Deficiency – 440 35.1.9 Hypoxanthine-Guanine Phosphoribosyltransferase Deficiency – 441 35.1.10 Adenine Phosphoribosyltransferase Deficiency – 442 35.1.11 Deoxyguanosine Kinase Deficiency – 442 35.2 Inborn Errors of Pyrimidine Metabolism – 445 35.2.1 UMP Synthase Deficiency (Hereditary Orotic Aciduria) – 445 35.2.2 Dihydropyrimidine Dehydrogenase Deficiency – 445 35.2.3 Dihydropyrimidinase Deficiency – 446 35.2.4 Ureidopropionase Deficiency – 446 35.2.5 Pyrimidine 5’-Nucleotidase Deficiency – 446 35.2.6 Cytosolic 5’-Nucleotidase Superactivity – 447 35.2.7 Thymidine Phosphorylase Deficiency – 447 35.2.8 Thymidine Kinase Deficiency – 447 References – 447 434 Chapter 35 · Disorders of Purine and Pyrimidine Metabolism Purine Metabolism Purine nucleotides are essential cellular constituents 4 The catabolic pathway starts from GMP, IMP and which intervene in energy transfer, metabolic regula- AMP, and produces uric acid, a poorly soluble tion, and synthesis of DNA and RNA. Purine metabo- compound, which tends to crystallize once its lism can be divided into three pathways: plasma concentration surpasses 6.5–7 mg/dl (0.38– 4 The biosynthetic pathway, often termed de novo, 0.47 mmol/l). starts with the formation of phosphoribosyl pyro- 4 The salvage pathway utilizes the purine bases, gua- phosphate (PRPP) and leads to the synthesis of nine, hypoxanthine and adenine, which are pro- inosine monophosphate (IMP). -

Chuanxiong Rhizoma Compound on HIF-VEGF Pathway and Cerebral Ischemia-Reperfusion Injury’S Biological Network Based on Systematic Pharmacology
ORIGINAL RESEARCH published: 25 June 2021 doi: 10.3389/fphar.2021.601846 Exploring the Regulatory Mechanism of Hedysarum Multijugum Maxim.-Chuanxiong Rhizoma Compound on HIF-VEGF Pathway and Cerebral Ischemia-Reperfusion Injury’s Biological Network Based on Systematic Pharmacology Kailin Yang 1†, Liuting Zeng 1†, Anqi Ge 2†, Yi Chen 1†, Shanshan Wang 1†, Xiaofei Zhu 1,3† and Jinwen Ge 1,4* Edited by: 1 Takashi Sato, Key Laboratory of Hunan Province for Integrated Traditional Chinese and Western Medicine on Prevention and Treatment of 2 Tokyo University of Pharmacy and Life Cardio-Cerebral Diseases, Hunan University of Chinese Medicine, Changsha, China, Galactophore Department, The First 3 Sciences, Japan Hospital of Hunan University of Chinese Medicine, Changsha, China, School of Graduate, Central South University, Changsha, China, 4Shaoyang University, Shaoyang, China Reviewed by: Hui Zhao, Capital Medical University, China Background: Clinical research found that Hedysarum Multijugum Maxim.-Chuanxiong Maria Luisa Del Moral, fi University of Jaén, Spain Rhizoma Compound (HCC) has de nite curative effect on cerebral ischemic diseases, *Correspondence: such as ischemic stroke and cerebral ischemia-reperfusion injury (CIR). However, its Jinwen Ge mechanism for treating cerebral ischemia is still not fully explained. [email protected] †These authors share first authorship Methods: The traditional Chinese medicine related database were utilized to obtain the components of HCC. The Pharmmapper were used to predict HCC’s potential targets. Specialty section: The CIR genes were obtained from Genecards and OMIM and the protein-protein This article was submitted to interaction (PPI) data of HCC’s targets and IS genes were obtained from String Ethnopharmacology, a section of the journal database. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -
Type of the Paper (Article
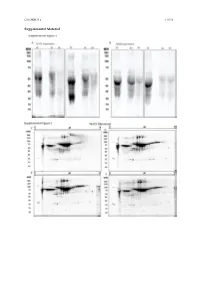
Cells 2020, 9, x 1 of 19 Supplemental Material Cells 2020, 9, x 2 of 19 Figure S1. Secretome enrichment: protocol optimization. 1D SDS-PAGE documentation of washing steps: Culture medium was substituted with FCS-free medium, which was changed every 2 h. The supernatants were then collected, and the proteins isolated and separated in 1D SDS-PAGE ((A) TK173 and (B) TK188). Proteins were stained with Flamingo fluorescent gel stain. Two-dimensional pattern of the proteins isolated from supernatant of TK173, (C) 2 h, (D) 4 h, (E) 6 h, and (F) 8 h after changing to FCS-free medium. (G) Cell secretome collected 24 h after elimination of the contaminating FCS-proteins with different washing steps. Proteins were stained with Flamingo fluorescent gel stain. Cells 2020, 9, x 3 of 19 Figure S2. 2-DE reference maps of secretomes; 150 μg proteins were loaded on an 11 cm IPG strip with a linear pH gradient PI 5–8 for IEF; 12% SDS-polyacrylamide gels were used for the second dimension. Proteins were stained with Flamingo fluorescent gel stain. Identified spots were assigned a number corresponding to that in their table. 2-DE maps from secretome of (A) TK173 control and (B) TGFβ1- treated ones. The 2-DE patterns revealed an alteration of secretome in stimulated TK173. Secretome patterns from TK173 treated with (C) ANG II and (D) PDGF. Cells 2020, 9, x 4 of 19 A Figure S3. Classification of the differentially expressed proteins upon ANG II, TGFβ1, or PDGF treatment in TK173. (A) Bar charts of the cellular component analyzed by STRAP biological function analysis in which the identified proteins from all treatments in both cell types are involved. -

Supplementary Methods
Supplementary methods Human lung tissues and tissue microarray (TMA) All human tissues were obtained from the Lung Cancer Specialized Program of Research Excellence (SPORE) Tissue Bank at the M.D. Anderson Cancer Center (Houston, TX). A collection of 26 lung adenocarcinomas and 24 non-tumoral paired tissues were snap-frozen and preserved in liquid nitrogen for total RNA extraction. For each tissue sample, the percentage of malignant tissue was calculated and the cellular composition of specimens was determined by histological examination (I.I.W.) following Hematoxylin-Eosin (H&E) staining. All malignant samples retained contained more than 50% tumor cells. Specimens resected from NSCLC stages I-IV patients who had no prior chemotherapy or radiotherapy were used for TMA analysis by immunohistochemistry. Patients who had smoked at least 100 cigarettes in their lifetime were defined as smokers. Samples were fixed in formalin, embedded in paraffin, stained with H&E, and reviewed by an experienced pathologist (I.I.W.). The 413 tissue specimens collected from 283 patients included 62 normal bronchial epithelia, 61 bronchial hyperplasias (Hyp), 15 squamous metaplasias (SqM), 9 squamous dysplasias (Dys), 26 carcinomas in situ (CIS), as well as 98 squamous cell carcinomas (SCC) and 141 adenocarcinomas. Normal bronchial epithelia, hyperplasia, squamous metaplasia, dysplasia, CIS, and SCC were considered to represent different steps in the development of SCCs. All tumors and lesions were classified according to the World Health Organization (WHO) 2004 criteria. The TMAs were prepared with a manual tissue arrayer (Advanced Tissue Arrayer ATA100, Chemicon International, Temecula, CA) using 1-mm-diameter cores in triplicate for tumors and 1.5 to 2-mm cores for normal epithelial and premalignant lesions. -

Supplementary Table S1 List of Proteins Identified with LC-MS/MS in the Exudates of Ustilaginoidea Virens Mol
Supplementary Table S1 List of proteins identified with LC-MS/MS in the exudates of Ustilaginoidea virens Mol. weight NO a Protein IDs b Protein names c Score d Cov f MS/MS Peptide sequence g [kDa] e Succinate dehydrogenase [ubiquinone] 1 KDB17818.1 6.282 30.486 4.1 TGPMILDALVR iron-sulfur subunit, mitochondrial 2 KDB18023.1 3-ketoacyl-CoA thiolase, peroxisomal 6.2998 43.626 2.1 ALDLAGISR 3 KDB12646.1 ATP phosphoribosyltransferase 25.709 34.047 17.6 AIDTVVQSTAVLVQSR EIALVMDELSR SSTNTDMVDLIASR VGASDILVLDIHNTR 4 KDB11684.1 Bifunctional purine biosynthetic protein ADE1 22.54 86.534 4.5 GLAHITGGGLIENVPR SLLPVLGEIK TVGESLLTPTR 5 KDB16707.1 Proteasomal ubiquitin receptor ADRM1 12.204 42.367 4.3 GSGSGGAGPDATGGDVR 6 KDB15928.1 Cytochrome b2, mitochondrial 34.9 58.379 9.4 EFDPVHPSDTLR GVQTVEDVLR MLTGADVAQHSDAK SGIEVLAETMPVLR 7 KDB12275.1 Aspartate 1-decarboxylase 11.724 112.62 3.6 GLILTLSEIPEASK TAAIAGLGSGNIIGIPVDNAAR 8 KDB15972.1 Glucosidase 2 subunit beta 7.3902 64.984 3.2 IDPLSPQQLLPASGLAPGR AAGLALGALDDRPLDGR AIPIEVLPLAAPDVLAR AVDDHLLPSYR GGGACLLQEK 9 KDB15004.1 Ribose-5-phosphate isomerase 70.089 32.491 32.6 GPAFHAR KLIAVADSR LIAVADSR MTFFPTGSQSK YVGIGSGSTVVHVVDAIASK 10 KDB18474.1 D-arabinitol dehydrogenase 1 19.425 25.025 19.2 ENPEAQFDQLKK ILEDAIHYVR NLNWVDATLLEPASCACHGLEK 11 KDB18473.1 D-arabinitol dehydrogenase 1 11.481 10.294 36.6 FPLIPGHETVGVIAAVGK VAADNSELCNECFYCR 12 KDB15780.1 Cyanovirin-N homolog 85.42 11.188 31.7 QVINLDER TASNVQLQGSQLTAELATLSGEPR GAATAAHEAYK IELELEK KEEGDSTEKPAEETK LGGELTVDER NATDVAQTDLTPTHPIR 13 KDB14501.1 14-3-3 -

Integrative Analysis of Promising Molecular Biomarkers and Pathways for Coronary Artery Disease Using WGCNA and Metade Methods
MOLECULAR MEDICINE REPORTS 18: 2789-2797, 2018 Integrative analysis of promising molecular biomarkers and pathways for coronary artery disease using WGCNA and MetaDE methods SHILIN YAN Department of Cardiology, Yangling Demonstration Zone Hospital, Xianyang, Shaanxi 712100, P.R. China Received January 8, 2018; Accepted May 31, 2018 DOI: 10.3892/mmr.2018.9277 Abstract. The present study aimed to examine the molecular ‘Fc gamma R‑mediated phagocytosis’ pathways may serve mechanisms of coronary artery disease (CAD). A total of four important roles in CAD. microarray datasets (training dataset no. GSE12288; valida- tion dataset nos. GSE20680, GSE20681 and GSE42148) were Introduction downloaded from the Gene Expression Omnibus database, which included CAD and healthy samples. Weighted gene Coronary artery disease (CAD) remains one of the most co-expression network analysis was applied to identify highly common causes of morbidity and mortality globally, with preserved modules across the four datasets. Differentially an increased prevalence predicted in the near future (1). expressed genes (DEGs) with significant consistency in the The occurrence of CAD events is primarily due to crosstalk four datasets were selected using the MetaDE method. The between genetic and environmental factors (2). A number of overlapping genes amongst the DEGs with significant consis- risk factors of CAD have been identified, including smoking, tency and in the preserved modules were used to construct obesity and family history (3). Available treatment options a protein-protein interaction (PPI) network, followed by primarily include lifestyle alterations, medical treatment functional enrichment analysis. A total of 11 modules were and surgical interventions, which are chosen depending on established in the training dataset, and five of them were highly comorbidities and the preferences of the individual patient (4). -

Nucleo'de)Metabolism) (Chapter)23)) INTRODUCTION)TO)NUCLEIC)ACIDS)
BIOCH 765: Biochemistry II Spring 2014 Nucleo'de)Metabolism) (Chapter)23)) Jianhan)Chen) Office)Hour:)MF)1:30B2:30PM,)Chalmers)034) Email:)[email protected]) Office:)785B2518) INTRODUCTION)TO)NUCLEIC)ACIDS) (c))Jianhan)Chen) 2) Eukaryo'c)cell) 3 Genome)is)believed)to)define)a)species) 4 Nucleic)Acids) • Polymer(of( nucleotides:(highly( flexible((compared( to(peptides)( • Nucleic(acids(are( universal(in(living( things,(as(they(are( found(in(all(cells( and(viruses.( • Nucleic(acids(were( first(discovered(by( Friedrich(Miescher( in(1871.( (c))Jianhan)Chen) 5 Building)Blocks)of)Nucleo'des) • Phosphate(group(+(pentose(carbohydrate(+( base((nitrogenGcontaining(heterocyclic(ring)( • Deoxyribonucleic(acid((DNA):(A/G/C/T( • Ribonuclei(acid((RNA):(A/G/C/U( Nucleotide nucleobase 6 Nucleo'des)have)many)roles) • Building(blocks(of(the(nucleic(acid(polymers(RNA(and( DNA.( • Energy(transfer(or(energy(coupling(to(drive(biosynthesis( and(other(processes((muscle(contraction,(transport,(etc).( • Oxidation(reduction(reactions.( • Intracellular(signaling.( ATP 7 Pentose)sugars) Base Phosphate sugar • The(pentose(sugar(in(the(nucloetide(is(either(ribose(or( deoxyribose.( ( – The(base(is(added(at(the(1(position(and(phosphates(are(added(at( the(5(position.( – Most(nucleotides,(including(those(incorporated(into(RNA,(contain( ribose.( – 2’GOH(reduced(to(–H(in(deoxynucleotides( – 3’GOH(participates(in(forming(phosphodiester(linkage( – Deoxynucleotides(are(exclusively(used(for(DNA(synthesis.( 8 Nucleo'de)forma'on) O ║ H N The base is added at the 1 6 N position and phosphates -

C6cc08797c1.Pdf
Electronic Supplementary Material (ESI) for Chemical Communications. This journal is © The Royal Society of Chemistry 2017 Supplementary Information for Competition-based, quantitative chemical proteomics in breast cancer cells identifies new target profiles for sulforaphane James A. Clulow,a,‡ Elisabeth M. Storck,a,‡ Thomas Lanyon-Hogg,a,‡ Karunakaran A. Kalesh,a Lyn Jonesb and Edward W. Tatea,* a Department of Chemistry, Imperial College London, London, UK, SW7 2AZ b Worldwide Medicinal Chemistry, Pfizer, 200 Cambridge Park Drive, MA 02140, USA ‡ Authors share equal contribution * corresponding author, email: [email protected] Table of Contents 1 Supporting Figures..........................................................................................................................3 2 Supporting Tables.........................................................................................................................17 2.1 Supporting Table S1. Incorporation validation for the R10K8 label in the 'spike-in' SILAC proteome of the MCF7 cell line........................................................................................................17 2.2 Supporting Table S2. Incorporation validation for the R10K8 label in the 'spike-in' SILAC proteome of the MDA-MB-231 cell line ...........................................................................................17 2.3 Supporting Table S3. High- and medium- confidence targets of sulforaphane in the MCF7 cell line..............................................................................................................................................17