Hurdles for Phage Therapy (PT) to Become a Reality
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Synthesis and Biological Evaluation of Trisindolyl-Cycloalkanes and Bis- Indolyl Naphthalene Small Molecules As Potent Antibacterial and Antifungal Agents
Synthesis and Biological Evaluation of Trisindolyl-Cycloalkanes and Bis- Indolyl Naphthalene Small Molecules as Potent Antibacterial and Antifungal Agents Dissertation Zur Erlangung des akademischen Grades doctor rerum naturalium (Dr. rer. nat.) Vorgelegt der Naturwissenschaftlichen Fakultät I Institut für Pharmazie Fachbereich für Pharmazeutische Chemie der Martin-Luther-Universität Halle-Wittenberg von Kaveh Yasrebi Geboren am 09.14.1987 in Teheran/Iran (Islamische Republik) Gutachter: 1. Prof. Dr. Andreas Hilgeroth (Martin-Luther-Universität Halle-Wittenberg, Germany) 2. Prof. Dr. Sibel Süzen (Ankara Üniversitesi, Turkey) 3. Prof. Dr. Michael Lalk (Ernst-Moritz-Arndt-Universität Greifswald, Germany) Halle (Saale), den 21. Juli 2020 Selbstständigkeitserklärung Hiermit erkläre ich gemäß § 5 (2) b der Promotionsordnung der Naturwissenschaftlichen Fakultät I – Institut für Pharmazie der Martin-Luther-Universität Halle-Wittenberg, dass ich die vorliegende Arbeit selbstständig und ohne Benutzung anderer als der angegebenen Hilfsmittel und Quellen angefertigt habe. Alle Stellen, die wörtlich oder sinngemäß aus Veröffentlichungen entnommen sind, habe ich als solche kenntlich gemacht. Ich erkläre ferner, dass diese Arbeit in gleicher oder ähnlicher Form bisher keiner anderen Prüfbehörde zur Erlangung des Doktorgrades vorgelegt wurde. Halle (Saale), den 21. Juli 2020 Kaveh Yasrebi Acknowledgement This study was carried out from June 2015 to July 2017 in the Research Group of Drug Development and Analysis led by Prof. Dr. Andreas Hilgeroth at the Institute of Pharmacy, Martin-Luther-Universität Halle-Wittenberg. I would like to thank all the people for their participation who supported my work in this way and helped me obtain good results. First of all, I would like to express my gratitude to Prof. Dr. Andreas Hilgeroth for providing me with opportunity to carry out my Ph.D. -

Novel Antimicrobial Agents Inhibiting Lipid II Incorporation Into Peptidoglycan Essay MBB
27 -7-2019 Novel antimicrobial agents inhibiting lipid II incorporation into peptidoglycan Essay MBB Mark Nijland S3265978 Supervisor: Prof. Dr. Dirk-Jan Scheffers Molecular Microbiology University of Groningen Content Abstract..............................................................................................................................................2 1.0 Peptidoglycan biosynthesis of bacteria ........................................................................................3 2.0 Novel antimicrobial agents ...........................................................................................................4 2.1 Teixobactin ...............................................................................................................................4 2.2 tridecaptin A1............................................................................................................................7 2.3 Malacidins ................................................................................................................................8 2.4 Humimycins ..............................................................................................................................9 2.5 LysM ........................................................................................................................................ 10 3.0 Concluding remarks .................................................................................................................... 11 4.0 references ................................................................................................................................. -
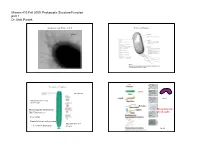
Parsek Micro410 Lecture
Microm 410 Fall 2009: Prokaryotic Structure/Function part 1 Dr. Matt Parsek Organization of the Prokaryotic Cell Prokaryotic Structures fimbriae Size Range of Prokaryotes bacillus See Table 4.1 (rigid) vibrio Nanobacteria 0.05‐0.2 µm (0.14‐0.2 µm) (flexible) Thiomargarita namibiensis Mycoplasma are 700‐750 µm (Fig. 4.2) pleomorphic green alga Nanochlorum eukaryotum Mycoplasma 0.1‐ ~1-2 µm in diameter 0.3 µm Fig. 4.1 Microm 410 Fall 2009: Prokaryotic Structure/Function part 1 Dr. Matt Parsek Staining cells for Microscopic observation Cell Arrangements Motility- ~80% of prokaryotes are motile streptococcus Staining properties: Gram Stain staphylococcus Fig. 2.3 Gram Stain (1884) (Bacteria) Gram-negative mixed culture Gram-positive Fig. 2.3 and 2.4 Microm 410 Fall 2009: Prokaryotic Structure/Function part 1 Dr. Matt Parsek Functions of the cytoplasmic membrane The phospholipid bi‐layer Fig. 4.9 Fig. 4.4 What is the structure of bacterial phospholipids? Other components of the cytoplasmic membrane Figs. 4.5‐4.6 Microm 410 Fall 2009: Prokaryotic Structure/Function part 1 Dr. Matt Parsek Archaeal membranes can be a lipid monolayer Archaeal phospholipids have an ether linkage Fig. 4.7 Fig. 4.8 Importance of Cell Wall Schematic diagram cell wall • Provides rigidity to cell allowing cell to withstand the large osmotic/ionic Fig. 4.16 changes a bacterium may experience in its environment, and turgor pressure of cytoplasm (conc. of solutes in cytoplasm). Cell lysis • May have a role in shape determination. • Provides a barrier against certain toxic chemical and biological agents. • Site of action of some of the most commonly used antibiotics used to treat bacterial infections (penicillin family). -
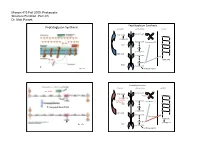
Parsek Lecture #3
Microm 410 Fall 2009: Prokaryotic Structure/Function: Part 2/3 Dr. Matt Parsek Peptidoglycan Synthesis Peptidoglycan Synthesis cytoplasm cell membrane cell wall Bactoprenol-P Pi UDP-NAM M G pentapeptide G M Bactoprenol Bactoprenol-P-P P M UMP G P NAM G M pentapeptide M G UDP-NAG Bactoprenol G P NAM‐NAG P NAM-NAG UMP pentapeptide Fig. 6.7a Interbridge peptide Peptidoglycan Synthesis Cross-linking of Peptidoglycan Strands cytoplasm cell membrane cell wall autolysins Bactoprenol-P Pi UDP-NAM Bacitracin M G pentapeptide G D-cycloserine Bactoprenol M (Oxamycin) Bactoprenol-P-P P M UMP G P Transpeptidase (FtsI) NAM G M pentapeptide M G UDP-NAG Bactoprenol G Vancomycin P NAM‐NAG P NAM-NAG pentapeptide UMP Fig. 6.7b pentapeptide Interbridge peptide Microm 410 Fall 2009: Prokaryotic Structure/Function: Part 2/3 Dr. Matt Parsek Cross-linking of Peptidoglycan Strands Antibiotic Resistance autolysins • Inactivate antibiotic β-lactamase (penicillinase) Clavulanic acid β-lactams Augmentin and Trimentin (combination of clavulanic acid and transpeptidase amoxicillin or ampicillin respectively) penicillins and cephalosporins lysozyme • Change chemistry of target site • Limit access of the antibiotic to target site Fig. 6.5 Cell Shape Determination • Modifications made to Peptidoglycan: ‐ lysozyme: Protoplasts/spheroplasts ‐ autolysins Bacillus subtilis ‐ endopeptidase Heliobacter pylori • Protein(s) may play a major role ‐ MreB protein Caulobacter crescentus ‐ MreB has homology to actin, a component of the cytoskeleton of eukaryotes. Shape determining protein‐ crescentin Fig. 6.4 Microm 410 Fall 2009: Prokaryotic Structure/Function: Part 2/3 Dr. Matt Parsek Cell Wall Gram-positive Bacteria intermediate filaments in the bacteria Caulobacter crescentus glycerol similar predicted structures of crescentin and intermediate filaments Fig. -

WO 2015/028850 Al 5 March 2015 (05.03.2015) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2015/028850 Al 5 March 2015 (05.03.2015) P O P C T (51) International Patent Classification: AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, C07D 519/00 (2006.01) A61P 39/00 (2006.01) BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, C07D 487/04 (2006.01) A61P 35/00 (2006.01) DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, A61K 31/5517 (2006.01) A61P 37/00 (2006.01) HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KN, KP, KR, A61K 47/48 (2006.01) KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, (21) International Application Number: OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, PCT/IB2013/058229 SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, (22) International Filing Date: TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, 2 September 2013 (02.09.2013) ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (26) Publication Language: English GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, SZ, TZ, (71) Applicant: HANGZHOU DAC BIOTECH CO., LTD UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, [US/CN]; Room B2001-B2019, Building 2, No 452 Sixth TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, Street, Hangzhou Economy Development Area, Hangzhou EE, ES, FI, FR, GB, GR, HR, HU, IE, IS, IT, LT, LU, LV, City, Zhejiang 310018 (CN). -

Genomic Analysis of the Recent Viral Isolate Vb Bthp-Goe4 Reveals Increased Diversity of Φ29-Like Phages
viruses Article Genomic Analysis of the Recent Viral Isolate vB_BthP-Goe4 Reveals Increased Diversity of φ29-Like Phages Tobias Schilling 1, Michael Hoppert 2 and Robert Hertel 1,* 1 Department of Genomic and Applied Microbiology & Göttingen Genomics Laboratory, Institute of Microbiology and Genetics, Georg-August-University Göttingen, 37077 Göttingen, Germany; [email protected] 2 Department of General Microbiology, Institute of Microbiology and Genetics, Georg-August-University Göttingen, 37077 Göttingen, Germany; [email protected] * Correspondence: [email protected]; Tel.: +49-551-39-91120 Received: 19 October 2018; Accepted: 8 November 2018; Published: 13 November 2018 Abstract: We present the recently isolated virus vB_BthP-Goe4 infecting Bacillus thuringiensis HD1. Morphological investigation via transmission electron microscopy revealed key characteristics of the genus Phi29virus, but with an elongated head resulting in larger virion particles of approximately 50 nm width and 120 nm height. Genome sequencing and analysis resulted in a linear phage chromosome of approximately 26 kb, harbouring 40 protein-encoding genes and a packaging RNA. Sequence comparison confirmed the relation to the Phi29virus genus and genomes of other related strains. A global average nucleotide identity analysis of all identified φ29-like viruses revealed the formation of several new groups previously not observed. The largest group includes Goe4 and may significantly expand the genus Phi29virus (Salasvirus) or the Picovirinae subfamily. Keywords: Bacillus; thuringiensis; vB_BthP-Goe4; Goe4; Picovirinae; Phi29virus; Salasvirus; Luci; bacteriophage; phage; pRNA 1. Introduction Bacteriophages or phages are viruses of bacteria and probably the most common biological entities on earth. Phage species outnumber their hosts by 10 times [1] and thus, represent the largest unexplored genetic reservoir. -

Antibiotic Discovery
ANTIBIOTIC DISCOVERY RESISTANCE PROFILING OF MICROBIAL GENOMES TO REVEAL NOVEL ANTIBIOTIC NATURAL PRODUCTS By CHELSEA WALKER, H. BSc. A Thesis Submitted to the School of Graduate Studies in Partial Fulfilment of the Requirements for the Degree Master of Science McMaster University © Copyright by Chelsea Walker, May 2017 McMaster University MASTER OF SCIENCE (2017) Hamilton, Ontario (Biochemistry and Biomedical Sciences) TITLE: Resistance Profiling of Microbial Genomes to Reveal Novel Antibiotic Natural Products. AUTHOR: Chelsea Walker, H. BSc. (McMaster University) SUPERVISOR: Dr. Nathan A. Magarvey. COMMITTEE MEMBERS: Dr. Eric Brown and Dr. Michael G. Surette. NUMBER OF PAGES: xvii, 168 ii Lay Abstract It would be hard to imagine a world where we could no longer use the antibiotics we are routinely being prescribed for common bacterial infections. Currently, we are in an era where this thought could become a reality. Although we have been able to discover antibiotics in the past from soil dwelling microbes, this approach to discovery is being constantly challenged. At the same time, the bacteria are getting smarter in their ways to evade antibiotics, in the form of resistance, or self-protection mechanisms. As such is it essential to devise methods which can predict the potential for resistance to the antibiotics we use early in the discovery and isolation process. By using what we have learned in the past about how bacteria protect themselves for antibiotics, we can to stay one step ahead of them as we continue to search for new sources of antibiotics from bacteria. iii Abstract Microbial natural products have been an invaluable resource for providing clinically relevant therapeutics for almost a century, including most of the commonly used antibiotics that are still in medical use today. -

E. Coli Pbp1b, Moenomycin-Based
Investigating the Ligand Interactions Between E. coli PBP1b, Moenomycin-based Compounds, and Beta-Lactam Compounds Peter Alexander MSc by Research 2017 i CERTIFICATE OF ORIGINALITY This is to certify that I am responsible for the work submitted in this thesis, that the original work is my own, except as specified in the acknowledgements and in references, and that neither the thesis nor the original work contained therein has been previously submitted to any institution for a degree. Signature: Name: Date: CERTIFICATE OF COMPLIANCE This is to certify that this project has been carried out in accordance with University principles regarding ethics and health and safety. Forms are available to view on request. Signature: Name: Date: ii Abstract Antimicrobial resistance is a growing problem in this era. Resistance to the majority of clinical antibiotics including those of a ‘last line of defence’ nature has been seen in a number of laboratory and clinical settings. One method aiming at reducing this problem is altering existing antimicrobial compounds, in order to improve pharmacological effects (avoiding resistance mechanisms, improved spectrum of use). Analysis of the interactions between the antimicrobial compounds and their targets can determine whether modifications to current antimicrobials (such as moenomycin A, a glycosyltransferase inhibitor) have altered the mode of action. ecoPBP1B is a bifunctional glycosyltransferase that could be used as a model for beta lactams and moenomycins, aiding in the design and development of novel antimicrobials based on these families. Moenomycin A has not seen high clinical usage due to poor pharmacokinetics and bioavailability. This project aimed to show whether ecoPBP1b can be used as a model for novel antimicrobials, such as seeing whether Moenomycin A analogues (with cell penetrating peptides to facilitate entry into the bacterial cell) still retain their ability to bind to glycosyltransferases. -

TESIS DOCTORAL Estudio Metagenómico De La Comunidad De
TESIS DOCTORAL Estudio metagenómico de la comunidad de virus y de su interacción con la microbiota en la cavidad bucal humana Marcos Parras Moltó Madrid, 2019 Estudio metagenómico de la comunidad de virus y de su interacción con la microbiota en la cavidad bucal humana Memoria presentada por Marcos Parras Moltó para optar al título de Doctor por la Universidad Autónoma de Madrid Esta Tesis se ha realizado en el Centro de Biología Molecular Severo Ochoa bajo la supervisión del Tutor y Director Alberto López Bueno, en el Programa de Doctorado en Biociencias Moleculares (RD 99/2011) Universidad Autónoma de Madrid Facultad de Ciencias Departamento de Biología Molecular Centro de Biología Molecular Severo Ochoa (CBMSO) Madrid, 2019 El Dr. Alberto López Bueno, Profesor Contratado Doctor en el Departamento de Biología Molecular de la Universidad Autónoma de Madrid (UAM) e investigador en el Centro de Biología Molecular Severo Ochoa (CBMSO): CERTIFICA: Haber dirigido y supervisado la Tesis Doctoral titulada "Estudio metagenómico de la comunidad de virus y de su interacción con la microbiota en la cavidad bucal humana” realizada por D. Marcos Parras Moltó, en el Programa de Doctorado en Biociencias Moleculares de la Universidad Autónoma de Madrid, por lo que autoriza la presentación de la misma. Madrid, a 23 de Abril de 2019, Alberto López Bueno La presente tesis doctoral ha sido posible gracias a la concesión de una “Ayuda para Contratos Predoctorales para la Formación de Doctores” convocatoria de 2013 (BES-2013-064773) asociada al proyecto SAF2012-38421 del Ministerio de Economía y Competitividad. Durante esta tesis se realizó una estancia de dos meses en el laboratorio del Catedrático Francisco Rodríguez Valera, director de grupo de investigación: Evolutionary Genomics Group de la Universidad Miguel Hernández de Elche (San Juan de Alicante), gracias a una “Ayuda a la Movilidad Predoctoral para la Realización de Estancias Breves en Centros de I+D” convocatoria de 2015 (EEBB-I-16-11876) concedida por el Ministerio de Economía y Competitividad. -

New Tools for Viral Metagenome Comparison and Assembled Virome Analysis Simon Roux1,2, Jeremy Tournayre1,2, Antoine Mahul3, Didier Debroas1,2 and François Enault1,2*
Roux et al. BMC Bioinformatics 2014, 15:76 http://www.biomedcentral.com/1471-2105/15/76 SOFTWARE Open Access Metavir 2: new tools for viral metagenome comparison and assembled virome analysis Simon Roux1,2, Jeremy Tournayre1,2, Antoine Mahul3, Didier Debroas1,2 and François Enault1,2* Abstract Background: Metagenomics, based on culture-independent sequencing, is a well-fitted approach to provide insights into the composition, structure and dynamics of environmental viral communities. Following recent advances in sequencing technologies, new challenges arise for existing bioinformatic tools dedicated to viral metagenome (i.e. virome) analysis as (i) the number of viromes is rapidly growing and (ii) large genomic fragments can now be obtained by assembling the huge amount of sequence data generated for each metagenome. Results: To face these challenges, a new version of Metavir was developed. First, all Metavir tools have been adapted to support comparative analysis of viromes in order to improve the analysis of multiple datasets. In addition to the sequence comparison previously provided, viromes can now be compared through their k-mer frequencies, their taxonomic compositions, recruitment plots and phylogenetic trees containing sequences from different datasets. Second, a new section has been specifically designed to handle assembled viromes made of thousands of large genomic fragments (i.e. contigs). This section includes an annotation pipeline for uploaded viral contigs (gene prediction, similarity search against reference viral genomes and protein domains) and an extensive comparison between contigs and reference genomes. Contigs and their annotations can be explored on the website through specifically developed dynamic genomic maps and interactive networks. Conclusions: The new features of Metavir 2 allow users to explore and analyze viromes composed of raw reads or assembled fragments through a set of adapted tools and a user-friendly interface. -

Lipid II: a Central Component in Bacterial Cell Wall Synthesis and a Target for Antibiotics
ARTICLE IN PRESS Prostaglandins, Leukotrienes and Essential Fatty Acids 79 (2008) 117–121 Contents lists available at ScienceDirect Prostaglandins, Leukotrienes and Essential Fatty Acids journal homepage: www.elsevier.com/locate/plefa Lipid II: A central component in bacterial cell wall synthesis and a target for antibiotics Ben de Kruijff Ã, Vincent van Dam, Eefjan Breukink Chemical Biology and Organic Chemistry, Utrecht University, Padualaan 8, Utrecht, The Netherlands abstract The bacterial cell wall is mainly composed of peptidoglycan, which is a three-dimensional network of long aminosugar strands located on the exterior of the cytoplasmic membrane. These strands consist of alternating MurNAc and GlcNAc units and are interlinked to each other via peptide moieties that are attached to the MurNAc residues. Peptidoglycan subunits are assembled on the cytoplasmic side of the bacterial membrane on a polyisoprenoid anchor and one of the key components in the synthesis of peptidoglycan is Lipid II. Being essential for bacterial cell survival, it forms an attractive target for antibacterial compounds such as vancomycin and several lantibiotics. Lipid II consists of one GlcNAc- MurNAc-pentapeptide subunit linked to a polyiosoprenoid anchor 11 subunits long via a pyrophosphate linker. This review focuses on this special molecule and addresses three questions. First, why are special lipid carriers as polyprenols used in the assembly of peptidoglycan? Secondly, how is Lipid II translocated across the bacterial cytoplasmic membrane? And finally, how is Lipid II used as a receptor for lantibiotics to kill bacteria? & 2008 Elsevier Ltd. All rights reserved. 1. Introduction which will be discussed later. Despite considerable knowledge of cell wall synthesis several key questions remained unanswered so The bacterial cell wall is a unique structure. -

Bacteriophage Therapy for Application Against Staphylococcus Aureus Infection and Biofilm in Chronic Rhinosinusitis
Bacteriophage therapy for application against Staphylococcus aureus infection and biofilm in chronic rhinosinusitis Amanda Jane Drilling Faculty of Health Sciences School of Medicine Discipline of Surgery March 2015 The enemy of my enemy is my friend i Table of Contents I. Abstract ....................................................................................................................................... v II. Declaration ................................................................................................................................ vii III. Acknowledgments ................................................................................................................ viii IV. Presentations and Awards Arising from this thesis ................................................................. x V. List of tables .............................................................................................................................. xii VI. List of Figures ...................................................................................................................... xiii VII. Abbreviations ......................................................................................................................... 1 1 Systematic review of the Literature ........................................................................................... 3 1.1 Rhinosinusitis ..................................................................................................................... 3 1.1.1 Acute and Chronic