Antibiotic Discovery
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Synthesis and Biological Evaluation of Trisindolyl-Cycloalkanes and Bis- Indolyl Naphthalene Small Molecules As Potent Antibacterial and Antifungal Agents
Synthesis and Biological Evaluation of Trisindolyl-Cycloalkanes and Bis- Indolyl Naphthalene Small Molecules as Potent Antibacterial and Antifungal Agents Dissertation Zur Erlangung des akademischen Grades doctor rerum naturalium (Dr. rer. nat.) Vorgelegt der Naturwissenschaftlichen Fakultät I Institut für Pharmazie Fachbereich für Pharmazeutische Chemie der Martin-Luther-Universität Halle-Wittenberg von Kaveh Yasrebi Geboren am 09.14.1987 in Teheran/Iran (Islamische Republik) Gutachter: 1. Prof. Dr. Andreas Hilgeroth (Martin-Luther-Universität Halle-Wittenberg, Germany) 2. Prof. Dr. Sibel Süzen (Ankara Üniversitesi, Turkey) 3. Prof. Dr. Michael Lalk (Ernst-Moritz-Arndt-Universität Greifswald, Germany) Halle (Saale), den 21. Juli 2020 Selbstständigkeitserklärung Hiermit erkläre ich gemäß § 5 (2) b der Promotionsordnung der Naturwissenschaftlichen Fakultät I – Institut für Pharmazie der Martin-Luther-Universität Halle-Wittenberg, dass ich die vorliegende Arbeit selbstständig und ohne Benutzung anderer als der angegebenen Hilfsmittel und Quellen angefertigt habe. Alle Stellen, die wörtlich oder sinngemäß aus Veröffentlichungen entnommen sind, habe ich als solche kenntlich gemacht. Ich erkläre ferner, dass diese Arbeit in gleicher oder ähnlicher Form bisher keiner anderen Prüfbehörde zur Erlangung des Doktorgrades vorgelegt wurde. Halle (Saale), den 21. Juli 2020 Kaveh Yasrebi Acknowledgement This study was carried out from June 2015 to July 2017 in the Research Group of Drug Development and Analysis led by Prof. Dr. Andreas Hilgeroth at the Institute of Pharmacy, Martin-Luther-Universität Halle-Wittenberg. I would like to thank all the people for their participation who supported my work in this way and helped me obtain good results. First of all, I would like to express my gratitude to Prof. Dr. Andreas Hilgeroth for providing me with opportunity to carry out my Ph.D. -
(12) United States Patent (10) Patent No.: US 7,879,798 B1 Aufseeser (45) Date of Patent: Feb
US007879798B1 (12) United States Patent (10) Patent No.: US 7,879,798 B1 Aufseeser (45) Date of Patent: Feb. 1, 2011 (54) COMPOSITION FOR INDOLENT WOUND Mazzotta, M.Y. “Nutrition and Wound Healing.” J. Am. Podiatr. Med. HEALING AND METHODS OF USE Assoc. 84(9): 456-462, Sep. 1994. THEREFOR Niedermeier, S. "Tierexperimentelle Untersuchungen Zur Frage der Behandlung von Hornhautlasionen Animal experiment studies on the problem of treating corneal lesions.” Klin. Monatsbl. (75) Inventor: Leslie S. Aufseeser, Lakewood, NJ (US) Augenheilkd (Germany, West), 190 (1):28-9, Jan. 1987. Seifter, E. etal. “Impaired Wound Healing in Streptozotocin diabetes. (73) Assignee: Regenicel, Inc., Lakewood, NJ (US) Prevention by supplemental Vitamin A.” Ann. Surgery (US). 194(1): 42-50, Jul. 1981. (*) Notice: Subject to any disclaimer, the term of this Gilmore, O.J.A. etal. “Aetiology and prevention of wound infection patent is extended or adjusted under 35 in appendicetomy.” British Journal of Surgery, 61:281-287. Mar. U.S.C. 154(b) by 594 days. 1974. Lowenfels, Albert B. “Viewpoints: Wound Healing With No Oint (21) Appl. No.: 11/895,474 ment, Non-antibiotic Ointment, or Antibiotic Ointment”. Medscape General Surgery. 8(2), 2006. (22) Filed: Aug. 24, 2007 United States Food and Drug Administration. “First Aid Antibiotic Drug Products”. Code of Federal Regulations, Title 21, vol. 5, Chap (51) Int. Cl. ter I, Part 333, Subpart B, pp. 222-225, Apr. 1, 2006. A6 IK 38/12 (2006.01) Akpek, EK et al. "A randomized trial of low-dose, topical mitomycin-C in the treatment of severe vernal keratoconjunctivitis.” (52) U.S. -

Novel Antimicrobial Agents Inhibiting Lipid II Incorporation Into Peptidoglycan Essay MBB
27 -7-2019 Novel antimicrobial agents inhibiting lipid II incorporation into peptidoglycan Essay MBB Mark Nijland S3265978 Supervisor: Prof. Dr. Dirk-Jan Scheffers Molecular Microbiology University of Groningen Content Abstract..............................................................................................................................................2 1.0 Peptidoglycan biosynthesis of bacteria ........................................................................................3 2.0 Novel antimicrobial agents ...........................................................................................................4 2.1 Teixobactin ...............................................................................................................................4 2.2 tridecaptin A1............................................................................................................................7 2.3 Malacidins ................................................................................................................................8 2.4 Humimycins ..............................................................................................................................9 2.5 LysM ........................................................................................................................................ 10 3.0 Concluding remarks .................................................................................................................... 11 4.0 references ................................................................................................................................. -
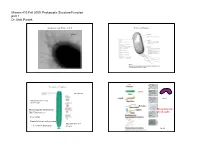
Parsek Micro410 Lecture
Microm 410 Fall 2009: Prokaryotic Structure/Function part 1 Dr. Matt Parsek Organization of the Prokaryotic Cell Prokaryotic Structures fimbriae Size Range of Prokaryotes bacillus See Table 4.1 (rigid) vibrio Nanobacteria 0.05‐0.2 µm (0.14‐0.2 µm) (flexible) Thiomargarita namibiensis Mycoplasma are 700‐750 µm (Fig. 4.2) pleomorphic green alga Nanochlorum eukaryotum Mycoplasma 0.1‐ ~1-2 µm in diameter 0.3 µm Fig. 4.1 Microm 410 Fall 2009: Prokaryotic Structure/Function part 1 Dr. Matt Parsek Staining cells for Microscopic observation Cell Arrangements Motility- ~80% of prokaryotes are motile streptococcus Staining properties: Gram Stain staphylococcus Fig. 2.3 Gram Stain (1884) (Bacteria) Gram-negative mixed culture Gram-positive Fig. 2.3 and 2.4 Microm 410 Fall 2009: Prokaryotic Structure/Function part 1 Dr. Matt Parsek Functions of the cytoplasmic membrane The phospholipid bi‐layer Fig. 4.9 Fig. 4.4 What is the structure of bacterial phospholipids? Other components of the cytoplasmic membrane Figs. 4.5‐4.6 Microm 410 Fall 2009: Prokaryotic Structure/Function part 1 Dr. Matt Parsek Archaeal membranes can be a lipid monolayer Archaeal phospholipids have an ether linkage Fig. 4.7 Fig. 4.8 Importance of Cell Wall Schematic diagram cell wall • Provides rigidity to cell allowing cell to withstand the large osmotic/ionic Fig. 4.16 changes a bacterium may experience in its environment, and turgor pressure of cytoplasm (conc. of solutes in cytoplasm). Cell lysis • May have a role in shape determination. • Provides a barrier against certain toxic chemical and biological agents. • Site of action of some of the most commonly used antibiotics used to treat bacterial infections (penicillin family). -
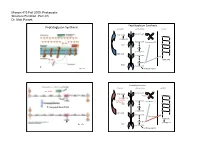
Parsek Lecture #3
Microm 410 Fall 2009: Prokaryotic Structure/Function: Part 2/3 Dr. Matt Parsek Peptidoglycan Synthesis Peptidoglycan Synthesis cytoplasm cell membrane cell wall Bactoprenol-P Pi UDP-NAM M G pentapeptide G M Bactoprenol Bactoprenol-P-P P M UMP G P NAM G M pentapeptide M G UDP-NAG Bactoprenol G P NAM‐NAG P NAM-NAG UMP pentapeptide Fig. 6.7a Interbridge peptide Peptidoglycan Synthesis Cross-linking of Peptidoglycan Strands cytoplasm cell membrane cell wall autolysins Bactoprenol-P Pi UDP-NAM Bacitracin M G pentapeptide G D-cycloserine Bactoprenol M (Oxamycin) Bactoprenol-P-P P M UMP G P Transpeptidase (FtsI) NAM G M pentapeptide M G UDP-NAG Bactoprenol G Vancomycin P NAM‐NAG P NAM-NAG pentapeptide UMP Fig. 6.7b pentapeptide Interbridge peptide Microm 410 Fall 2009: Prokaryotic Structure/Function: Part 2/3 Dr. Matt Parsek Cross-linking of Peptidoglycan Strands Antibiotic Resistance autolysins • Inactivate antibiotic β-lactamase (penicillinase) Clavulanic acid β-lactams Augmentin and Trimentin (combination of clavulanic acid and transpeptidase amoxicillin or ampicillin respectively) penicillins and cephalosporins lysozyme • Change chemistry of target site • Limit access of the antibiotic to target site Fig. 6.5 Cell Shape Determination • Modifications made to Peptidoglycan: ‐ lysozyme: Protoplasts/spheroplasts ‐ autolysins Bacillus subtilis ‐ endopeptidase Heliobacter pylori • Protein(s) may play a major role ‐ MreB protein Caulobacter crescentus ‐ MreB has homology to actin, a component of the cytoskeleton of eukaryotes. Shape determining protein‐ crescentin Fig. 6.4 Microm 410 Fall 2009: Prokaryotic Structure/Function: Part 2/3 Dr. Matt Parsek Cell Wall Gram-positive Bacteria intermediate filaments in the bacteria Caulobacter crescentus glycerol similar predicted structures of crescentin and intermediate filaments Fig. -

B-Lactams: Chemical Structure, Mode of Action and Mechanisms of Resistance
b-Lactams: chemical structure, mode of action and mechanisms of resistance Ru´ben Fernandes, Paula Amador and Cristina Prudeˆncio This synopsis summarizes the key chemical and bacteriological characteristics of b-lactams, penicillins, cephalosporins, carbanpenems, monobactams and others. Particular notice is given to first-generation to fifth-generation cephalosporins. This review also summarizes the main resistance mechanism to antibiotics, focusing particular attention to those conferring resistance to broad-spectrum cephalosporins by means of production of emerging cephalosporinases (extended-spectrum b-lactamases and AmpC b-lactamases), target alteration (penicillin-binding proteins from methicillin-resistant Staphylococcus aureus) and membrane transporters that pump b-lactams out of the bacterial cell. Keywords: b-lactams, chemical structure, mechanisms of resistance, mode of action Historical perspective Alexander Fleming first noticed the antibacterial nature of penicillin in 1928. When working with Antimicrobials must be understood as any kind of agent another bacteriological problem, Fleming observed with inhibitory or killing properties to a microorganism. a contaminated culture of Staphylococcus aureus with Antibiotic is a more restrictive term, which implies the the mold Penicillium notatum. Fleming remarkably saw natural source of the antimicrobial agent. Similarly, under- the potential of this unfortunate event. He dis- lying the term chemotherapeutic is the artificial origin of continued the work that he was dealing with and was an antimicrobial agent by chemical synthesis [1]. Initially, able to describe the compound around the mold antibiotics were considered as small molecular weight and isolates it. He named it penicillin and published organic molecules or metabolites used in response of his findings along with some applications of penicillin some microorganisms against others that inhabit the same [4]. -

WO 2015/028850 Al 5 March 2015 (05.03.2015) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2015/028850 Al 5 March 2015 (05.03.2015) P O P C T (51) International Patent Classification: AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, C07D 519/00 (2006.01) A61P 39/00 (2006.01) BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, C07D 487/04 (2006.01) A61P 35/00 (2006.01) DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, A61K 31/5517 (2006.01) A61P 37/00 (2006.01) HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KN, KP, KR, A61K 47/48 (2006.01) KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, (21) International Application Number: OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, PCT/IB2013/058229 SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, (22) International Filing Date: TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, 2 September 2013 (02.09.2013) ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (26) Publication Language: English GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, SZ, TZ, (71) Applicant: HANGZHOU DAC BIOTECH CO., LTD UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, [US/CN]; Room B2001-B2019, Building 2, No 452 Sixth TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, Street, Hangzhou Economy Development Area, Hangzhou EE, ES, FI, FR, GB, GR, HR, HU, IE, IS, IT, LT, LU, LV, City, Zhejiang 310018 (CN). -

Download (12Mb)
A Thesis Submitted for the Degree of PhD at the University of Warwick Permanent WRAP URL: http://wrap.warwick.ac.uk/110352 Copyright and reuse: This thesis is made available online and is protected by original copyright. Please scroll down to view the document itself. Please refer to the repository record for this item for information to help you to cite it. Our policy information is available from the repository home page. For more information, please contact the WRAP Team at: [email protected] warwick.ac.uk/lib-publications THE BRITISH LIBRARY BRITISH THESIS SERVICE THE DISTRIBUTION OF PHENOTYPIC AND GENOTYPIC CHARACTERS WITHIN STREPTOMYCETES AND THEIR RELATIONSHIP TITLE . TO ANTIBIOTIC PRODUCTION. AUTHOR........ Lesley Phillips, DEGREE.................................................... AWARDING BODY _ TI. .. The University of Warwick, THESIS NUMBER THIS THESIS HAS BEEN MICROFILMED EXACTLY AS RECEIVED The quality of this reproduction is dependent upon the quality of the original thesis submitted for microfilming. Every effort has been made to ensure the highest quality of reproduction. Some pages may have indistinct print, especially if the original papers were poorly produced or if the awarding body sent an inferior copy. If pages are missing, please contact the awarding body which granted the degree. Previously copyrighted materials (journal articles, published texts, etc.) are not filmed. This copy of the thesis has been supplied on condition that anyone who consults it is understood to recognise that Its copyright rests with its author and that no information derived from it may be published without the author's prior written consent. Reproduction of this thesis, other than as permitted under the United Kingdom Copyright Designs and Patents Act 1988, or under specific agreement with the copyright holder, is prohibited. -

From Sugar Based Bio Renewable Resources to New Chemical Building Blocks and Bioactive Molecules
Raquel Alexandra Germano Nunes Licenciatura em Química Aplicada From sugar based bio renewable resources to new chemical building blocks and bioactive molecules Dissertação para obtenção do Grau de Mestre em Química Bioorgânica Orientador: Prof. Doutor Carlos Alberto Mateus Afonso, FF-UL Co-orientador: Rafael Filipe Teixeira Arbuéz Gomes, MSc, FF-UL Presidente: Prof. Doutora Paula Cristina de Sério Branco, FCT-UNL Arguente: Prof. Doutora Luísa Maria da Silva Pinto Ferreira, FCT-UNL Vogal: Prof. Doutor Carlos Alberto Mateus Afonso, FF-UL Março 2017 LOMBADA Raquel Nunes Raquel From sugar based bio renewable resources to new chemical building blocks and bioactive molecules bioactive and blocks chemical new building to resources renewable bio based sugar From 2017 Raquel Alexandra Germano Nunes Licenciatura em Química Aplicada From sugar based bio renewable resources to new chemical building blocks and bioactive molecules Dissertação para obtenção do Grau de Mestre em Química Bioorgânica Orientador: Prof. Doutor Carlos Alberto Mateus Afonso, FF-UL Co-orientador: Rafael Filipe Teixeira Arbuéz Gomes, MSc, FF-UL Presidente: Prof. Doutora Paula Cristina de Sério Branco, FCT-UNL Arguente: Prof. Doutora Luísa Maria da Silva Pinto Ferreira, FCT-UNL Vogal: Prof. Doutor Carlos Alberto Mateus Afonso, FF-UL Março 2017 From sugar based bio renewable resources to new chemical building blocks and bioactive molecules Copyright © Raquel Alexandra Germano Nunes, Faculdade de Ciências e Tecnologia, Universidade Nova de Lisboa. A Faculdade de Ciências e Tecnologia e a Universidade Nova de Lisboa têm o direito, perpétuo e sem limites geográficos, de arquivar e publicar esta dissertação através de exemplares impressos reproduzidos em papel ou de forma digital, ou por outro qualquer meio conhecido ou que venha a ser inventado e de divulgar através de repositórios científicos e de admitir a sua cópia e distribuição com objectivos educacionais ou de investigação, não comerciais, desde que seja dado crédito ao autor e editor. -

General Prescription
GENERAL PRESCRIPTION LESSON 1. INTRODUCTION. PRESCRIPTION. SOLID MEDICINAL FORMS Objective: To study the structure of the prescription, learn the rules and get practical skills in writing out solid medicinal forms in prescription. To carry out practical tasks on prescriptions it is recommended to use Appendix 1. Key questions: 1. Pharmacology as a science and the basis of therapy. Main development milestones of modern pharmacology. Sections of Pharmacology. 2. The concept of medicinal substance, medicinal agent (medicinal drug, drug), medicinal form. 3. The concept of the pharmacological action and types of the action of drugs. 4. The sources of obtaining drugs. 5. International and national pharmacopeia, its content and purpose. 6. Pharmacy. Rules of drug storage and dispensing. 7. Prescription and its structure. Prescription forms. General rules for writing out a prescription. State regulation of writing out and dispensing drugs. 8. Name of medicinal products (international non-proprietary name - INN, trade name). 9. Peculiarities of writing out narcotic, poisonous and potent substances in prescription. 10. Drugs under control. Drugs prohibited for prescribing. 11. Solid medicinal forms: tablets, dragee (pills), powders, capsules. Their characteristics, advantages and disadvantages. Rules of prescribing. Write out prescriptions for: 1. 5 powders of Codeine 0.015 g. 1 powder orally twice a day. 2. 10 powders of Didanosine 0.25 g in sachets to prepare solution for internal use. Accept inside twice a day one sachet powder after dissolution in a glass of boiled water. 3. 50 mg of Alteplase powder in the bottle. Dissolve the content of the bottle in 50 ml of saline. First 15 ml introduce intravenously streamly, then intravenous drip. -

Anew Drug Design Strategy in the Liht of Molecular Hybridization Concept
www.ijcrt.org © 2020 IJCRT | Volume 8, Issue 12 December 2020 | ISSN: 2320-2882 “Drug Design strategy and chemical process maximization in the light of Molecular Hybridization Concept.” Subhasis Basu, Ph D Registration No: VB 1198 of 2018-2019. Department Of Chemistry, Visva-Bharati University A Draft Thesis is submitted for the partial fulfilment of PhD in Chemistry Thesis/Degree proceeding. DECLARATION I Certify that a. The Work contained in this thesis is original and has been done by me under the guidance of my supervisor. b. The work has not been submitted to any other Institute for any degree or diploma. c. I have followed the guidelines provided by the Institute in preparing the thesis. d. I have conformed to the norms and guidelines given in the Ethical Code of Conduct of the Institute. e. Whenever I have used materials (data, theoretical analysis, figures and text) from other sources, I have given due credit to them by citing them in the text of the thesis and giving their details in the references. Further, I have taken permission from the copyright owners of the sources, whenever necessary. IJCRT2012039 International Journal of Creative Research Thoughts (IJCRT) www.ijcrt.org 284 www.ijcrt.org © 2020 IJCRT | Volume 8, Issue 12 December 2020 | ISSN: 2320-2882 f. Whenever I have quoted written materials from other sources I have put them under quotation marks and given due credit to the sources by citing them and giving required details in the references. (Subhasis Basu) ACKNOWLEDGEMENT This preface is to extend an appreciation to all those individuals who with their generous co- operation guided us in every aspect to make this design and drawing successful. -

E. Coli Pbp1b, Moenomycin-Based
Investigating the Ligand Interactions Between E. coli PBP1b, Moenomycin-based Compounds, and Beta-Lactam Compounds Peter Alexander MSc by Research 2017 i CERTIFICATE OF ORIGINALITY This is to certify that I am responsible for the work submitted in this thesis, that the original work is my own, except as specified in the acknowledgements and in references, and that neither the thesis nor the original work contained therein has been previously submitted to any institution for a degree. Signature: Name: Date: CERTIFICATE OF COMPLIANCE This is to certify that this project has been carried out in accordance with University principles regarding ethics and health and safety. Forms are available to view on request. Signature: Name: Date: ii Abstract Antimicrobial resistance is a growing problem in this era. Resistance to the majority of clinical antibiotics including those of a ‘last line of defence’ nature has been seen in a number of laboratory and clinical settings. One method aiming at reducing this problem is altering existing antimicrobial compounds, in order to improve pharmacological effects (avoiding resistance mechanisms, improved spectrum of use). Analysis of the interactions between the antimicrobial compounds and their targets can determine whether modifications to current antimicrobials (such as moenomycin A, a glycosyltransferase inhibitor) have altered the mode of action. ecoPBP1B is a bifunctional glycosyltransferase that could be used as a model for beta lactams and moenomycins, aiding in the design and development of novel antimicrobials based on these families. Moenomycin A has not seen high clinical usage due to poor pharmacokinetics and bioavailability. This project aimed to show whether ecoPBP1b can be used as a model for novel antimicrobials, such as seeing whether Moenomycin A analogues (with cell penetrating peptides to facilitate entry into the bacterial cell) still retain their ability to bind to glycosyltransferases.