Download The
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Quarantine Regulation for Importation of Plants
Quarantine Requirements for The Importation of Plants or Plant Products into The Republic of China Bureau of Animal and Plant Health Inspection and Quarantine Council of Agriculture Executive Yuan In case of any discrepancy between the Chinese text and the English translation thereof, the Chinese text shall govern. Updated November 26, 2009 更新日期:2009 年 12 月 16 日 - 1 - Quarantine Requirements for The Importation of Plants or Plant Products into The Republic of China A. Prohibited Plants or Plant Products Pursuant to Paragraph 1, Article 14, Plant Protection and Quarantine Act 1. List of prohibited plants or plant products, countries or districts of origin and the reasons for prohibition: Plants or Plant Products Countries or Districts of Origin Reasons for Prohibition 1. Entire or any part of the All countries and districts 1. Rice hoja blanca virus following living plants (Tenuivirus) (excluding seeds): 2. Rice dwarf virus (1) Brachiaria spp. (Phytoreovirus) (2) Echinochloa spp. 3. Rice stem nematode (3) Panicum spp. (Ditylenchus angustus (4) Paspalum spp. Butler) (5) Oryza spp., Leersia hexandra, Saccioleps interrupta (6) Rottboellia spp. (7) Triticum aestivum 2. Entire or any part of the Asia and Pacific Region West Indian sweet potato following living plants (1) Palau weevil (excluding seeds) (2) China (Euscepes postfasciatus (1) Calystegia spp. (3) Cook Islands Fairmaire) (2) Dioscorea japonica (4) Federated States of Micronesia (3) Ipomoea spp. (5) Fiji (4) Pharbitis spp. (6) Guam (7) Kiribati (8) New Caledonia (9) Norfolk Island -

Ventura County Plant Species of Local Concern
Checklist of Ventura County Rare Plants (Twenty-second Edition) CNPS, Rare Plant Program David L. Magney Checklist of Ventura County Rare Plants1 By David L. Magney California Native Plant Society, Rare Plant Program, Locally Rare Project Updated 4 January 2017 Ventura County is located in southern California, USA, along the east edge of the Pacific Ocean. The coastal portion occurs along the south and southwestern quarter of the County. Ventura County is bounded by Santa Barbara County on the west, Kern County on the north, Los Angeles County on the east, and the Pacific Ocean generally on the south (Figure 1, General Location Map of Ventura County). Ventura County extends north to 34.9014ºN latitude at the northwest corner of the County. The County extends westward at Rincon Creek to 119.47991ºW longitude, and eastward to 118.63233ºW longitude at the west end of the San Fernando Valley just north of Chatsworth Reservoir. The mainland portion of the County reaches southward to 34.04567ºN latitude between Solromar and Sequit Point west of Malibu. When including Anacapa and San Nicolas Islands, the southernmost extent of the County occurs at 33.21ºN latitude and the westernmost extent at 119.58ºW longitude, on the south side and west sides of San Nicolas Island, respectively. Ventura County occupies 480,996 hectares [ha] (1,188,562 acres [ac]) or 4,810 square kilometers [sq. km] (1,857 sq. miles [mi]), which includes Anacapa and San Nicolas Islands. The mainland portion of the county is 474,852 ha (1,173,380 ac), or 4,748 sq. -

NBPGR Okf"Kzd Izfrosnu ANNUAL REPORT 2012-2013
ISSN NO 0971-2572 NBPGR okf"kZd izfrosnu ANNUAL REPORT 2012-2013 jk"Vªh; ikni vkuqOakf'kd Laklk/u C;wjks (Hkkjrh; Ñf"k vuqLak/ku ifj"kn) iwlk ifjlj] ubZ fnYyh&110 012 NATIONAL BUREAU OF PLANT GENETIC RESOURCES (Indian Council of Agricultural Research) Pusa Campus, New Delhi - 110 012 Citation : Anonymous (2013). Annual Report of the National Bureau of Plant Genetic Resources 2012-2013, NBPGR, Pusa Campus, New Delhi, India, 186+vi p. Compiled and Edited by : Dr. Arjun Lal, Principal Scientist Dr. (Mrs.) Kavita Gupta, Principal Scientist Dr. (Mrs.) Vandana Tyagi, Principal Scientist Dr. (Mrs.) Sangita Yadav, Senior Scientist This report includes unprocessed or semi-processed data, which would form the basis of scientific papers in due course. The material contained in the report therefore may not be made use of without the written permission of the Director, National Bureau of Plant Genetic Resources, New Delhi except for quoting it for scientific reference. Published by the Director, National Bureau of Plant Genetic Resources, Pusa Campus, New Delhi-110 012, and Printed at Alpha Printographics (India), New Delhi-110 028. Tel.: 9999039940, 9811199620 CONTENTS Preface Executive Summary 1 Introduction 9 NBPGR Headquarters, New Delhi 1. Plant Exploration and Germplasm Collection 13 2. Germplasm Evaluation 19 3. Germplasm Conservation 40 4. Plant Quarantine 45 5. Germplasm Exchange 53 6. Tissue Culture and Cryopreservation 60 7. PGR Policy Planning 65 8. Agricultural Knowledge Management 67 9. Genomic Resources 70 NBPGR Regional Stations/ Base Centers 10. Regional Station, Akola 86 11. Regional Station, Bhowali 91 12. Base Center, Cuttack 96 13. -

JOURNAL of Agriculturalv RESEARCH
Vol.37 AUGUST 1,1928 No. 3 JOURNAL OF AGRICULTURAlv RESEARCH CONTENTS Pfige Hosts and Symptoms of Ring Spot, a Virus Disease of Plants - - - - - 127 S. A. WINGARD Bacterial Pocket Disease of the Sugar Beet - --- - - - - 155 NELLIE A. BROWN Growth and Senescence in Red Danish Cows as Measured by the Rate of Milk Secretion -----_--»-.... 1^9 W. L. GAINES and D. D, SHAW Morphology and Taxonomy of the Pecan-Scab Fungus, Cladosporium effusum (Wint.) Comb. Nov. -------.»-.... 18X J, B. DEMAREE PUBLISHED BY AUTHORITY OF THE SECRETARY OF AGRICULTURE WITH THE COOPERATION OF THE ASSOCIATION OF LAPÍD-GRANT COLLEGES AND UNIVERSITIES UNITED STATES GOVERNMENT PRINTING OFFICE WASHINGTON 1928 JOINT COMMITTEE ON POLICY AND MANUSCRIPTS FOR THE UNITED STATES DEPARTMENT FOR THE ASSOCIATION OF LAND-GRANT OF AGRICULTURE COLLEGES AND UNIVERSITIES B. W. ALLEN, CHAIRMAN R. W. THATCHER Chief, Office (^ Experiment Stations President^ Massachusetts Agricultural College C. L. SHEAR U. J. FUNCHESS Senior Pathologist in Charge, Mycology and Directort Alabama Experiment Station Disease Survey A. C. BAKER G. A. DEAN Senior Entomologist in Charge, Tropical and Head, Department of Entomology, Kansas Subtropical Plant Insect Investigations Agricultural Experiment Station EDITORUL SUPERVISION M. C. MERRILL Editorial Chief of Publications, United States Departmeni of Agriculture All correspondence regarding articles from State experiment stations should be addressed to R. W. Thatcher, Agricultural College, Amherst, Mass. Published on the first and fifteenth of each month. This volume will consist of twelve numbers and the Contents and Index. Subscription price: Domestic, $4.00 a year (two volumes) Single numbers, 20-cents Foreign, $5.00 a year (two volumes) Single numbers, 25 cents If separates are desired in quantity, they should be ordered at the time the manuscript is sent to the printer, and they will be supplied practically at cost. -

Genomic Characterization of the Cacao Swollen Shoot Virus Complex and Other Theobroma Cacao-Infecting Badnaviruses
Genomic Characterization of the Cacao Swollen Shoot Virus Complex and other Theobroma Cacao-Infecting Badnaviruses Item Type text; Electronic Dissertation Authors Chingandu, Nomatter Publisher The University of Arizona. Rights Copyright © is held by the author. Digital access to this material is made possible by the University Libraries, University of Arizona. Further transmission, reproduction or presentation (such as public display or performance) of protected items is prohibited except with permission of the author. Download date 29/09/2021 07:25:04 Link to Item http://hdl.handle.net/10150/621859 GENOMIC CHARACTERIZATION OF THE CACAO SWOLLEN SHOOT VIRUS COMPLEX AND OTHER THEOBROMA CACAO-INFECTING BADNAVIRUSES by Nomatter Chingandu __________________________ A Dissertation Submitted to the Faculty of the SCHOOL OF PLANT SCIENCES In Partial Fulfillment of the Requirements For the Degree of DOCTOR OF PHILOSOPHY WITH A MAJOR IN PLANT PATHOLOGY In the Graduate College THE UNIVERSITY OF ARIZONA 2016 1 THE UNIVERSITY OF ARIZONA GRADUATE COLLEGE As members of the Dissertation Committee, we certify that we have read the dissertation prepared by Nomatter Chingandu, entitled “Genomic characterization of the Cacao swollen shoot virus complex and other Theobroma cacao-infecting badnaviruses” and recommend that it be accepted as fulfilling the dissertation requirement for the Degree of Doctor of Philosophy. _______________________________________________________ Date: 7.27.2016 Dr. Judith K. Brown _______________________________________________________ Date: 7.27.2016 Dr. Zhongguo Xiong _______________________________________________________ Date: 7.27.2016 Dr. Peter J. Cotty _______________________________________________________ Date: 7.27.2016 Dr. Barry M. Pryor _______________________________________________________ Date: 7.27.2016 Dr. Marc J. Orbach Final approval and acceptance of this dissertation is contingent upon the candidate’s submission of the final copies of the dissertation to the Graduate College. -

DISEASE RESISTANCE TESTING in COCOA a Review on Behalf of FAO/INGENIC J.C. Zadoks, Consultant
Zadoks - Disease resistance in cocoa 1997 February 2, 1997 DISEASE RESISTANCE TESTING IN COCOA A review on behalf of FAO/INGENIC J.C. Zadoks, consultant do Department of Phytopathology P.O.Box 8025 6700 EE Wageningen The Netherlands private fax + 31 - 317 - 48 34 12 fax + 31 - 317 - 42 36 14 e-mail Jan Carel [email protected] [email protected] - (hoi o Zadoks - Disease resistance in cocoa 1997 TABLE OF CONTENTS 0. Executive summary 1. Introduction 1.1. INGENIC 1.2. Objectives of the review 1.3. Procedure 2. Background information. The host plant 2.1. Origin 2.2. Agronomy 2.3. Genetics 2.4. Breeding 2.5. Pre-breeding 2.6. Conclusions and recommendations 3. Background information. The pathogens 3.1. Taxonom ic cons iderations 3.2. Subspecific variation 3.3. Selected pathogens 3.4. General comments 4. Resistance testing 4.1. Resistance, tolerance and escape 4.2. Components of resistance 4.3. Host-parasite interactions 4.4. Methods of resistance testing 4.5. Predictive tests 4.6. Resistance tests in campo 4.7. Resistance tests in vivo 4.8. Biochemical and molecular tests 4.9. International aspects of resistance testing 5. Resistance 5.1. Past and present situation of resistance breeding 5.2. Breeding objectives 5.3. Results with selected pathogens 5.4. Molecular techniques 5.5. General comments 5.6. Conclusions 2 Zadoks - Disease resistance in cocoa 1997 6. International cooperation 6.1. Organizational aspects 6.2. Needs 6.3. Pre-breeding 6.4. Preliminaries to implementation 6.5. Quality standards 6.6. Recommendations 7. -

International Plant Genetic Resources Instit., (IPGRI)
FAO/IPGRINo. 20. Cacao Technical Guidelines for the Safe Movement of Germplasm No. 201 Cacao edited by E.A. Frison, M. Diekman and D. Nowell netic t Ge Res lan ou P rc al e n s o I ti n a s t n i r t u e t t e n I IPGRI in collaboration with the American Cocoa Research Institute No. 20. Cacao 2 Previously published Technical Guidelines for the Safe Movement of Germplasm These guidelines describe technical procedures that minimize the rist of pest introduc- tions with movement of germplasm for research, crop improvement, plant breeding, exploration or conservation. The recommendations and information in these guide- lines are intended for germplasm for research, conservation and plant breeding programmes. Recommendations for commercial consignments are not the objective of these guidelines. Cocoa 1989 Edible aroids 1989 Musa (1st edition) 1989 Sweet potato 1989 Yam 1989 Legumes 1990 Cassava 1991 Citrus 1991 Grapevine 1991 Vanilla 1991 Coconut 1993 Sugarcane 1993 Small fruits (Fragaria, Ribes, Rubus, Vaccinium) 1994 Musa spp. (2nd edition) 1996 Stone Fruits 1996 Eucalyptus spp. 1996 Allium 1998 Potato 1998 No. 20. Cacao 3 CONTENTS Introduction ................................................ 4 Contributors ............................................... 6 Intermediate quarantine ........................... 8 Stations available for cacao ................ 8 General recommendations ....................... 9 Options for the movement of cacao germplasm in relation to the risk of moving pests ............................................ 10 Descriptions of pests ............................... 12 Viruses ................................................. 12 Cacao necrosis nepovirus ............. 12 Cacao swollen shoot badnavirus . 13 Cacao yellow mosaic lymovirus.. 16 Other virus-like diseases ............ 16 Fungi .................................................... 17 Witches’ broom ............................. 17 Moniliophthora pod rot ................. 19 Vascular streak dieback ............... 21 Phytophthora spp. -

Ajay Kumar Tiwari Editor Advances in Seed Production and Management Advances in Seed Production and Management Ajay Kumar Tiwari Editor
Ajay Kumar Tiwari Editor Advances in Seed Production and Management Advances in Seed Production and Management Ajay Kumar Tiwari Editor Advances in Seed Production and Management Editor Ajay Kumar Tiwari UP Council of Sugarcane Research Shahjahanpur, Uttar Pradesh, India ISBN 978-981-15-4197-1 ISBN 978-981-15-4198-8 (eBook) https://doi.org/10.1007/978-981-15-4198-8 # Springer Nature Singapore Pte Ltd. 2020 This work is subject to copyright. All rights are reserved by the Publisher, whether the whole or part of the material is concerned, specifically the rights of translation, reprinting, reuse of illustrations, recitation, broadcasting, reproduction on microfilms or in any other physical way, and transmission or information storage and retrieval, electronic adaptation, computer software, or by similar or dissimilar methodology now known or hereafter developed. The use of general descriptive names, registered names, trademarks, service marks, etc. in this publication does not imply, even in the absence of a specific statement, that such names are exempt from the relevant protective laws and regulations and therefore free for general use. The publisher, the authors, and the editors are safe to assume that the advice and information in this book are believed to be true and accurate at the date of publication. Neither the publisher nor the authors or the editors give a warranty, expressed or implied, with respect to the material contained herein or for any errors or omissions that may have been made. The publisher remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. This Springer imprint is published by the registered company Springer Nature Singapore Pte Ltd. -

List of Viruses Presenting at the Wild State a Biological Risk for Plants
December 2008 List of viruses presenting at the wild state a biological risk for plants CR Species 2 Abutilon mosaic virus 2 Abutilon yellows virus 2 Aconitum latent virus 2 African cassava mosaic virus 2 Ageratum yellow vein virus 2 Agropyron mosaic virus 2 Ahlum waterborne virus 2 Alfalfa cryptic virus 1 2 Alfalfa mosaic virus 2 Alsike clover vein mosaic virus 2 Alstroemeria mosaic virus 2 Amaranthus leaf mottle virus 2 American hop latent virus ( ← Hop American latent virus) 2 American plum line pattern virus 2 Anthoxanthum latent blanching virus 2 Anthriscus yellows virus 2 Apple chlorotic leaf spot virus 2 Apple mosaic virus 2 Apple stem grooving virus 2 Apple stem pitting virus 2 Arabis mosaic virus satellite RNA 2 Araujia mosaic virus 2 Arracacha virus A 2 Artichoke Italian latent virus 2 Artichoke latent virus 2 Artichoke mottled crinkle virus 2 Artichoke yellow ringspot virus 2 Asparagus virus 1 2 Asparagus virus 2 2 Avocado sunblotch viroid 2 Bajra streak virus 2 Bamboo mosaic virus 2 Banana bract mosaic virus 2 Banana bunchy top virus 2 Banana streak virus 2 Barley mild mosaic virus page 1 December 2008 2 Barley mosaic virus 3 Barley stripe mosaic virus 2 Barley yellow dwarf virus-GPV 2 Barley yellow dwarf virus-MAV 2 Barley yellow dwarf virus-PAV 2 Barley yellow dwarf virus-RGV 2 Barley yellow dwarf virus-RMV 2 Barley yellow dwarf virus-SGV 2 Barley yellow mosaic virus 2 Barley yellow streak mosaic virus 2 Barley yellow striate mosaic virus 2 Bean calico mosaic virus 2 Bean common mosaic necrosis virus 2 Bean common mosaic -

Shared Flora of the Alta and Baja California Pacific Islands
Monographs of the Western North American Naturalist Volume 7 8th California Islands Symposium Article 12 9-25-2014 Island specialists: shared flora of the Alta and Baja California Pacific slI ands Sarah E. Ratay University of California, Los Angeles, [email protected] Sula E. Vanderplank Botanical Research Institute of Texas, 1700 University Dr., Fort Worth, TX, [email protected] Benjamin T. Wilder University of California, Riverside, CA, [email protected] Follow this and additional works at: https://scholarsarchive.byu.edu/mwnan Recommended Citation Ratay, Sarah E.; Vanderplank, Sula E.; and Wilder, Benjamin T. (2014) "Island specialists: shared flora of the Alta and Baja California Pacific slI ands," Monographs of the Western North American Naturalist: Vol. 7 , Article 12. Available at: https://scholarsarchive.byu.edu/mwnan/vol7/iss1/12 This Monograph is brought to you for free and open access by the Western North American Naturalist Publications at BYU ScholarsArchive. It has been accepted for inclusion in Monographs of the Western North American Naturalist by an authorized editor of BYU ScholarsArchive. For more information, please contact [email protected], [email protected]. Monographs of the Western North American Naturalist 7, © 2014, pp. 161–220 ISLAND SPECIALISTS: SHARED FLORA OF THE ALTA AND BAJA CALIFORNIA PACIFIC ISLANDS Sarah E. Ratay1, Sula E. Vanderplank2, and Benjamin T. Wilder3 ABSTRACT.—The floristic connection between the mediterranean region of Baja California and the Pacific islands of Alta and Baja California provides insight into the history and origin of the California Floristic Province. We present updated species lists for all California Floristic Province islands and demonstrate the disjunct distributions of 26 taxa between the Baja California and the California Channel Islands. -
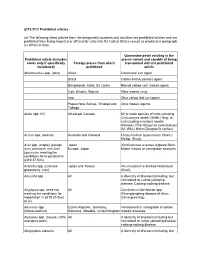
319.37-2 Prohibited Articles
§319.37-2 Prohibited articles. (a) The following listed articles from the designated countries and localities are prohibited articles and are prohibited from being imported or offered for entry into the United States except as provided in paragraph (c) of this section. Quarantine pests existing in the Prohibited article (includes places named and capable of being seeds only if specifically Foreign places from which transported with the prohibited mentioned) prohibited article Abelmoschus spp. (okra) Africa Cotton leaf curl agent. Brazil Cotton Anthocyanosis agent. Bangladesh, India, Sri Lanka Bhendi yellow vein mosaic agent. Cote d'Ivoire, Nigeria Okra mosaic virus. Iraq Okra yellow leaf curl agent. Papua New Guinea, Trinidad and Okra mosaic agents. Tobago Abies spp. (fir) All except Canada 50 or more species of rusts including Chrysomyxa abietis (Wallr.) Ung. (a rust causing a serious needle disease); Phacidiopycnis pseudotsuga (M. Wils.) Hahn (Douglas fir canker). Acacia spp. (acacia) Australia and Oceania Uromycladium tepperianum (Sacc.) McAlp. (Rust). Acer spp. (maple) (except Japan Xanthomonas acernea (Ogawa) Burk. Acer palmatum and Acer Europe, Japan Maple mosaic or variegation diseases. japonicum meeting the conditions for importation in §319.37-5(m) Actinidia spp. (Chinese Japan and Taiwan Pucciniastrum actinidiae Hiratusuka gooseberry, kiwi). (Rust). Adonidia spp All A diversity of diseases including, but not limited to: Lethal yellowing disease; Cadang-cadang disease. Aeglopsis spp. seed not All Candidatus Liberibacter spp. meeting the conditions for (Huanglongbing disease of citrus, importation in §319.37-5(w) Citrus greening). or (x). Aesculus spp. Czech Republic, Germany, Horsechestnut variegation or yellow (horsechestnut) Romania, Slovakia, United Kingdom mosaic diseases. -

An Update of the Host Range of Tomato Spotted Wilt Virus Giuseppe Parrella, Patrick Gognalons, Kahsay Gebre Selassie, C
An update of the host range of tomato spotted wilt virus Giuseppe Parrella, Patrick Gognalons, Kahsay Gebre Selassie, C. Vovlas, Georges Marchoux To cite this version: Giuseppe Parrella, Patrick Gognalons, Kahsay Gebre Selassie, C. Vovlas, Georges Marchoux. An update of the host range of tomato spotted wilt virus. Journal of Plant Pathology, Springer, 2003, 85 (4), pp.227-264. hal-02682821 HAL Id: hal-02682821 https://hal.inrae.fr/hal-02682821 Submitted on 1 Jun 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution - ShareAlike| 4.0 International License Journal of Plant Pathology (2003), 85 (4, Special issue), 227-264 Edizioni ETS Pisa, 2003 227 INVITED REVIEW AN UPDATE OF THE HOST RANGE OF TOMATO SPOTTED WILT VIRUS G. Parrella1, P. Gognalons2, K. Gebre-Selassiè2, C. Vovlas3 and G. Marchoux2 1 Istituto per la Protezione delle Piante del CNR, Sezione di Portici, Via Università 133, 80055 Portici (NA), Italy 2 Institute National de la Recherche Agronomique, Station de Pathologie Végétale, BP 94 - 84143 Montfavet Cedex, France 3 Dipartimento di Protezione delle Piante e Microbiologia Applicata, Università degli Studi and Istituto di Virologia Vegetale del CNR, Sezione di Bari, Via G.