Identification of Successful Mentoring Communities Using Network-Based
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

G.N Lewis, Research Style by Glenn Seaborg
Lawrence Berkeley National Laboratory Recent Work Title THE RESEARCH STYLE OF GILBERT N. LEWIS: ACIDS AND BASES Permalink https://escholarship.org/uc/item/9wn2k3xt Author Seaborg, G.T. Publication Date 1982-10-01 eScholarship.org Powered by the California Digital Library University of California o LBL-14785e 'd- Lawrence Berkeley Laboratory UNIVERSITY OF CALIFORN~IARECEIVED LAWRENCE . OCT '27 1982 LIBRARY AND DOCUMENTS SECTION Presented at the l83rd National Meeting of' the American Chemical Society, Las Vegas, NV, March 30, 1982 THE RESEARCH STYLE OF GILBERT N. LEWIS: ACIDS AND BASES Glenn T. Seaborg TWO~WEEK LOAN COpy October 1982 This is a Library Circulating Copy which may be borrowed for two weeks. Fora personal retention copy~ call '-- Tech. Info. Division~ Ext. 6782. &r I; -~ .-...1 (\ c-C' ~ \' \; Prepared for the U.S. Department of Energy under Contract DE-AC03-76SF00098 DISCLAIMER This document was prepared as an account of work sponsored by the United States Government. While this document is believed to contain correct information, neither the United States Government nor any agency thereof, nor the Regents of the Uriiversity of California, nor any of their employees, makes any warranty, express or implied, or assumes any legal responsibility for the accuracy, completeness, or usefulness of any information, apparatus, product, or process disclosed, or represents that its use would not infringe privately owned rights. Reference herein to any specific commercial product, process, or service by its trade name, trademark, manufacturer, or otherwise, does not necessarily constitute or imply its endorsement, recommendation, or favoring by the United States Government or any agency thereof, or the.Regents of the University of California. -

Radiocarbon After Four Decades RADIOCARBON R.E
Radiocarbon After Four Decades RADIOCARBON R.E. Taylor A. Long R.S. Kra Editors Radiocarbon After Four Decades An Interdisciplinary Perspective With 148 Illustrations Springer Science+Business Media, LLC R.E. Thylor Austin Long Department of Anthropology Department of Geosciences Institute of Geophysics and Planetary Physics The University of Arizona University of California, Riverside Thscon, AZ 85721 USA Riverside, CA 92521-0418 USA Renee S. Kra Department of Geosciences The University of Arizona Thscon, AZ 85721 USA Library of Congress Cataloging-in-Publication Data Radiocarbon after four decades: an interdisciplinary perspective / [editors], R.E. Taylor, Austin Long, Renee S. Kra. p. cm. Includes bibliographical references and index. ISBN 978-1-4757-4251-0 ISBN 978-1-4757-4249-7 (eBook) DOI 1O.l007/978-1-4757-4249-7 1. Radiocarbon dating. I. Taylor, R. E. (Royal Ervin), 1938- II. Long, Austin. III. Kra, Renee S. QC798.D3R3 1992 546'.6815884-dc20 91-44448 Printed on acid-free paper. © 1992 Springer Science+BusinessMedia New York Originally published by Springer-Verlag New York, Inc. in 1992 Softcover reprint of the hardcover 1st edition 1992 All rights reserved. This work may not be translated or copied in whole or in part without the written per mission of the publisher,Springer Science+Bnsiness Media, LLC, except for brief excerpts in connection with reviews or scholarly analysis. Use in connection with any form of information storage and retrieval, electronic adaptation, computer software, or by similar or dissimilar methodology now known or hereaf ter developed is forbidden. The use of general descriptive names, trade names, trademarks, etc., in this publication, even if the former are not especially identified, is not to be taken as a sign that such names, as understood by the Trade Marks and Merchandise Marks Act, may accordingly be used freely by anyone. -
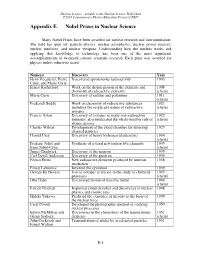
Appendix E Nobel Prizes in Nuclear Science
Nuclear Science—A Guide to the Nuclear Science Wall Chart ©2018 Contemporary Physics Education Project (CPEP) Appendix E Nobel Prizes in Nuclear Science Many Nobel Prizes have been awarded for nuclear research and instrumentation. The field has spun off: particle physics, nuclear astrophysics, nuclear power reactors, nuclear medicine, and nuclear weapons. Understanding how the nucleus works and applying that knowledge to technology has been one of the most significant accomplishments of twentieth century scientific research. Each prize was awarded for physics unless otherwise noted. Name(s) Discovery Year Henri Becquerel, Pierre Discovered spontaneous radioactivity 1903 Curie, and Marie Curie Ernest Rutherford Work on the disintegration of the elements and 1908 chemistry of radioactive elements (chem) Marie Curie Discovery of radium and polonium 1911 (chem) Frederick Soddy Work on chemistry of radioactive substances 1921 including the origin and nature of radioactive (chem) isotopes Francis Aston Discovery of isotopes in many non-radioactive 1922 elements, also enunciated the whole-number rule of (chem) atomic masses Charles Wilson Development of the cloud chamber for detecting 1927 charged particles Harold Urey Discovery of heavy hydrogen (deuterium) 1934 (chem) Frederic Joliot and Synthesis of several new radioactive elements 1935 Irene Joliot-Curie (chem) James Chadwick Discovery of the neutron 1935 Carl David Anderson Discovery of the positron 1936 Enrico Fermi New radioactive elements produced by neutron 1938 irradiation Ernest Lawrence -

UC San Diego UC San Diego Electronic Theses and Dissertations
UC San Diego UC San Diego Electronic Theses and Dissertations Title The new prophet : Harold C. Urey, scientist, atheist, and defender of religion Permalink https://escholarship.org/uc/item/3j80v92j Author Shindell, Matthew Benjamin Publication Date 2011 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California UNIVERSITY OF CALIFORNIA, SAN DIEGO The New Prophet: Harold C. Urey, Scientist, Atheist, and Defender of Religion A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in History (Science Studies) by Matthew Benjamin Shindell Committee in charge: Professor Naomi Oreskes, Chair Professor Robert Edelman Professor Martha Lampland Professor Charles Thorpe Professor Robert Westman 2011 Copyright Matthew Benjamin Shindell, 2011 All rights reserved. The Dissertation of Matthew Benjamin Shindell is approved, and it is acceptable in quality and form for publication on microfilm and electronically: ___________________________________________________________________ ___________________________________________________________________ ___________________________________________________________________ ___________________________________________________________________ ___________________________________________________________________ Chair University of California, San Diego 2011 iii TABLE OF CONTENTS Signature Page……………………………………………………………………...... iii Table of Contents……………………………………………………………………. iv Acknowledgements…………………………………………………………………. -

Thin Films Based on Gadolinium Applied to the Magnetic Refrigeration Asmaa Endichi
Thin films based on Gadolinium applied to the magnetic refrigeration Asmaa Endichi To cite this version: Asmaa Endichi. Thin films based on Gadolinium applied to the magnetic refrigeration. Physics [physics]. Université de Lorraine; Université Mohammed V (Rabat), 2020. English. NNT : 2020LORR0150. tel-03128459 HAL Id: tel-03128459 https://hal.univ-lorraine.fr/tel-03128459 Submitted on 2 Feb 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. AVERTISSEMENT Ce document est le fruit d'un long travail approuvé par le jury de soutenance et mis à disposition de l'ensemble de la communauté universitaire élargie. Il est soumis à la propriété intellectuelle de l'auteur. Ceci implique une obligation de citation et de référencement lors de l’utilisation de ce document. D'autre part, toute contrefaçon, plagiat, reproduction illicite encourt une poursuite pénale. Contact : [email protected] LIENS Code de la Propriété Intellectuelle. articles L 122. 4 Code de la Propriété Intellectuelle. articles L 335.2- L 335.10 http://www.cfcopies.com/V2/leg/leg_droi.php http://www.culture.gouv.fr/culture/infos-pratiques/droits/protection.htm THESE Pour l'obtention du titre de : DOCTEUR de L’UNIVERSITE DE LORRAINE Spécialité : Physique Présenté par : Asmaa ENDICHI Thin films based on Gadolinium applied to the magnetic refrigeration. -

Figuring out a Date Archaeologists Cannot Always Immediately Give a Date for Some Things They Find
Figuring Out a Date Archaeologists cannot always immediately give a date for some things they find. One way archaeologists try to figure out dates is by comparing kinds of materials used to make things. Early people used stone tools. Eventually, people discovered metal, which was better than stone, and they began using that. Archaeologists know that stone was used before metal because they find stone in deeper layers in the stratigraphy. How the ax developed: 1. The first axes were held in the hand. 2. Polished stone axes were used by the earliest farmers to clear land. 3. The first bronze axes copied the stone ones 4. Bronze axes developed to fit in wooden handles. 5. A new development used a loop to hold the ax to the handle. 6. A strong modern ax has a steel head. In the 1940s, there was a revolution in archaeology. An American chemist named Willard Libby discovered a new method for dating objects from the distant past, called radiocarbon, or carbon-14 dating. It is based on the scientific principle that all living things contain a certain amount of radioactive carbon. Once a living thing dies, carbon begins to decay. Scientists know that half of the carbon decays in 5,730 years. Measuring how much radioactive carbon is present in a sample gives a date. This kind of dating can be used for rocks, pottery, and glass. Charcoal is always a good sample for radiocarbon dating. 'r'*~neuti6ts. nitrogen and fonn 'Th~'heutrons . radioactive carbon ;interact with (C14). ",-.' ... Plants take in this ... In living plants .. -

The Links of Chain of Development of Physics from Past to the Present in a Chronological Order Starting from Thales of Miletus
ISSN (Online) 2393-8021 IARJSET ISSN (Print) 2394-1588 International Advanced Research Journal in Science, Engineering and Technology Vol. 5, Issue 10, October 2018 The Links of Chain of Development of Physics from Past to the Present in a Chronological Order Starting from Thales of Miletus Dr.(Prof.) V.C.A NAIR* Educational Physicist, Research Guide for Physics at Shri J.J.T. University, Rajasthan-333001, India. *[email protected] Abstract: The Research Paper consists mainly of the birth dates of scientists and philosophers Before Christ (BC) and After Death (AD) starting from Thales of Miletus with a brief description of their work and contribution to the development of Physics. The author has taken up some 400 odd scientists and put them in a chronological order. Nobel laureates are considered separately in the same paper. Along with the names of researchers are included few of the scientific events of importance. The entire chain forms a cascade and a ready reference for the reader. The graph at the end shows the recession in the earlier centuries and its transition to renaissance after the 12th century to the present. Keywords: As the contents of the paper mainly consists of names of scientists, the key words are many and hence the same is not given I. INTRODUCTION As the material for the topic is not readily available, it is taken from various sources and the collection and compiling is a Herculean task running into some 20 pages. It is given in 3 parts, Part I, Part II and Part III. In Part I the years are given in Chronological order as per the year of birth of the scientist and accordingly the serial number. -

08 Golf-Men Guide-Bleeds.Pmd
26 27 he San Francisco Bay Area is a major metropoli- tan area of approximately six million people and Tone of the most scenic regions in the United States. The Bay Area includes the major cities of San Francisco and Oakland, as well as Berkeley, home of the world- renowned University of California. Just south is the city of San Jose and the Silicon Valley, home to many of the world’s high-tech companies. The Bay Area also lies within easy driving distance of the high Sierra resorts of Lake Tahoe and Yosemite, the Monterey/Carmel penin- sula, the world famous Napa wine country, and the spec- tacular Mendocino Coast. Everyone knows “The City” - San Francisco - from count- less photographs, movies and television shows that cap- ture its magic. It is a city built on a series of more than 40 hills, offering panoramic views of every kind. The hub of a nine-county complex and the financial and insurance capi- tal of the world, San Francisco has a resident population of about 740,000. San Francisco is situated on a 46.6 square-mile peninsula bounded on the west by the Pa- cific Ocean, on the north by the Golden Gate strait, and from north to east by the San Francisco Bay. The City has been named the world’s top city twice by readers of Conde Nast Traveller and the top U.S. city seven times since 1988. The San Francisco Bay is spanned by two landmarks, the Golden Gate and San Francisco-Oakland Bay bridges, and graced by four islands: Alcatraz, Angel, Yerba Buena and Treasure. -

The Glacial World According to Wally
THE GLACIAL WORLD ACCORDING TO WALLY W.S. BROECKER ELDIGO PRESS 2002 THE GLACIAL WORLD ACCORDING TO WALLY W.S. BROECKER LAMONT-DOHERTY EARTH OBSERVATORY OF COLUMBIA UNIVERSITY Palisades, NY 10964 September 1992 Second Revised Edition June 1995 Third Revised Edition August 2002 FOREWORD In 1990 I started a book on glacial climates. I planned four sections, one on indicators, one on clocks, one on records and finally, one on the physics of the system. After completing the first three of these, I got stuck in my attempt to come up with a conceptual view of what might be driving these changes. However, having taught a course on climate change for many years, I know that much of the information one needs is not available in textbook form. Instead, we all depend heavily on reprints. As this proves to be an inefficient route for students interested in learning the basics of our field, I have tried to capture many of these basics in the first three sections of my would-be book. As I realized that it would be several years before I completed this project, I thought it might be useful to the community if I were to make available the three largely completed sections. This was done in 1992. In preparation for use in a short course to be taught at the Geological Society of America in November 2002, I undertook an effort to update the first three sections and the references during the early part of 2002. I encourage purchasers of the book to make as many copies as they wish for use by students and colleagues. -

Case Studies from the Archival and Cultural Heritage Program in The
QUICK DESIGN GUIDE Prac4cal archives, persistent cultural memory: Case studies from the Archival and Cultural Heritage program in the Graduate QUICK TIPS (--THIS SECTION DOES NOT PRINT--) School of Library and Informaon Science at Dominican University (--THIS SECTION DOES NOT PRINT--) This PowerPoint template requires basic PowerPoint (version 2007 or newer) skills. Below is a list of commonly This PowerPoint 2007 template produces a 36”x60” asked questions specific to this template. professional poster. It will save you valuable time placing Edward J. Valauskas ([email protected]) If you are using an older version of PowerPoint some titles, subtitles, text, and graphics. Graduate School of Library and Informaon Science, Dominican University, River Forest, IL template features may not work properly. Use it to create your presentation. Then send it to Using the template PosterPresentations.com for premium quality, same day Introduc4on Experiences with photographic archives Experiences with musical archives Experiences with digital collec4ons Verifying the quality of your graphics affordable printing. Go to the VIEW menu and click on ZOOM to set your Archival and Cultural Heritage studies at GSLIS combine classroom Displaced persons (DP) photographic archives, Balzekas Museum of The Da! Project, creation of an archives for the Chicago post-punk, Creation of an open access scholarly journal, World Libraries (http:// preferred magnification. This template is at 50% the size experiences with rich, interactive opportunities with collections and Lithuanian Culture, Chicago. existential band Da! www.worlib.org) We provide a series of online tutorials that will guide you of the final poster. All text and graphics will be printed at institutions in the Chicago area and elsewhere. -

Contributions of Civilizations to International Prizes
CONTRIBUTIONS OF CIVILIZATIONS TO INTERNATIONAL PRIZES Split of Nobel prizes and Fields medals by civilization : PHYSICS .......................................................................................................................................................................... 1 CHEMISTRY .................................................................................................................................................................... 2 PHYSIOLOGY / MEDECINE .............................................................................................................................................. 3 LITERATURE ................................................................................................................................................................... 4 ECONOMY ...................................................................................................................................................................... 5 MATHEMATICS (Fields) .................................................................................................................................................. 5 PHYSICS Occidental / Judeo-christian (198) Alekseï Abrikossov / Zhores Alferov / Hannes Alfvén / Eric Allin Cornell / Luis Walter Alvarez / Carl David Anderson / Philip Warren Anderson / EdWard Victor Appleton / ArthUr Ashkin / John Bardeen / Barry C. Barish / Nikolay Basov / Henri BecqUerel / Johannes Georg Bednorz / Hans Bethe / Gerd Binnig / Patrick Blackett / Felix Bloch / Nicolaas Bloembergen -
Stirred and Shaken!
Stirred and shaken! CHEMISTRY NOBELS In the International Year of Chemistry, C Sivaram charts the Nobel prize winning achievements in the field, from work on synthesis of sugars to vitamin chemistry. his year is the International Year of TChemistry. Major achievements in chemistry are recognised every year by way of the Nobel Prize. The very first Nobel Prize in Chemistry awarded in 1901 went to Dutch chemist Jacobus Vant Hoff mainly for formulating laws of osmotic pressure in solutions. The 1902 prize went to Emil Fischer for work on sugar chemistry leading to synthe- sis of sugars and purines. He was a student of Adolf Von Bayer, who himself got the prize in 1905 for synthesis of organic dyes, including that of the indigo dye. This proved disastrous for the Indian indigo plant export, which was earlier the only source for the dye. Two other Fischers also got the chem- istry Nobel. One was Hans Fischer, who got it in 1930 for the constitution of chlorophyll, use of substituted porphyrins leading to eventual synthesis of haemin. In 1973, Ernst Otto Fischer shared the MIXING IT UP The very first Nobel Prize prize with Geoffrey Wilkinson for their in- in Chemistry was awarded in 1901. dependent work on organometallic ‘sand- wich’ compounds and revolutionising tran- the DNA double helix model, Perutz and sition metal chemistry with compound like Kendrew got the chemistry prize the same Ferrocene and those compounding rheni- year for their work on globular proteins, um and hydrogen or chromium with ben- thus making it an all-British affair! zene among others.