Crystal Structures of Ophiostoma Piceae Sterol Esterase: Structural Insights Into Activation Mechanism and Product Release
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

MIAMI UNIVERSITY the Graduate School Certificate for Approving The
MIAMI UNIVERSITY The Graduate School Certificate for Approving the Dissertation We hereby approve the Dissertation of Matthew F. Rouhier Candidate for the Degree: Doctor of Philosophy ___________________________________________________________ Director (Dr. Ann Hagerman) ___________________________________________________________ Reader (Dr.Chris Makaroff) ___________________________________________________________ Reader (Dr.Gary Lorigan) __________________________________________________________ Reader (Dr. Richard Taylor) _________________________________________________________ Graduate School Representative (Dr. Paul James) ! ABSTRACT CHARACTERIZATION OF YDR036C FROM Saccharomyces cerevisiae by Matthew F. Rouhier Beta-hydroxyisobutyryl-CoA (HIBYL-CoA) hydrolases are found ubiquitously in eukaryotes where they function in the catabolism of valine. A homologous enzyme (YDR036C) is also present in yeast where valine catabolism is distinctly different and does not require a HIBYL-CoA hydrolase. Like the other eukaryotic hydrolases, the yeast hydrolase is a member of the crotonase super-family which catalyzes various reactions using CoA thioester substrates. Crotonases typically function in fatty acid degradation which is peroxisomal in yeast while YDR036C is found strictly in mitochondria. Like other HIBYL-CoA hydrolases the yeast enzyme demonstrated activity toward beta-hydroxyacyl-CoAs, but unlike the other HIBYL-CoA hydrolases the yeast activity is greatest with beta-hydroxypropionyl-CoA (3-HP-CoA). Further characterization of the active site has determined that the preference of the hydrolases for 3-HP- CoA or HIBYL-CoA is dependent upon the residue at position 177. The glutamate at position 121 is responsible for the coordination with the beta-hydroxyl group and replacement with valine renders the enzyme non-specific for a hydroxyl group. Another unique feature to yeast hydrolases is the presence of a C-terminal tail of 80 amino acids, which when removed renders the enzyme inactive, suggesting that it may play a role in substrate binding. -

(10) Patent No.: US 8119385 B2
US008119385B2 (12) United States Patent (10) Patent No.: US 8,119,385 B2 Mathur et al. (45) Date of Patent: Feb. 21, 2012 (54) NUCLEICACIDS AND PROTEINS AND (52) U.S. Cl. ........................................ 435/212:530/350 METHODS FOR MAKING AND USING THEMI (58) Field of Classification Search ........................ None (75) Inventors: Eric J. Mathur, San Diego, CA (US); See application file for complete search history. Cathy Chang, San Diego, CA (US) (56) References Cited (73) Assignee: BP Corporation North America Inc., Houston, TX (US) OTHER PUBLICATIONS c Mount, Bioinformatics, Cold Spring Harbor Press, Cold Spring Har (*) Notice: Subject to any disclaimer, the term of this bor New York, 2001, pp. 382-393.* patent is extended or adjusted under 35 Spencer et al., “Whole-Genome Sequence Variation among Multiple U.S.C. 154(b) by 689 days. Isolates of Pseudomonas aeruginosa” J. Bacteriol. (2003) 185: 1316 1325. (21) Appl. No.: 11/817,403 Database Sequence GenBank Accession No. BZ569932 Dec. 17. 1-1. 2002. (22) PCT Fled: Mar. 3, 2006 Omiecinski et al., “Epoxide Hydrolase-Polymorphism and role in (86). PCT No.: PCT/US2OO6/OOT642 toxicology” Toxicol. Lett. (2000) 1.12: 365-370. S371 (c)(1), * cited by examiner (2), (4) Date: May 7, 2008 Primary Examiner — James Martinell (87) PCT Pub. No.: WO2006/096527 (74) Attorney, Agent, or Firm — Kalim S. Fuzail PCT Pub. Date: Sep. 14, 2006 (57) ABSTRACT (65) Prior Publication Data The invention provides polypeptides, including enzymes, structural proteins and binding proteins, polynucleotides US 201O/OO11456A1 Jan. 14, 2010 encoding these polypeptides, and methods of making and using these polynucleotides and polypeptides. -

Cholesterol Esterase from Porcine Pancreas
Cholesterol Esterase from porcine pancreas Catalog Number C9464 Storage Temperature –20 °C CAS RN 9026-00-0 Substrates: EC 3.1.1.13 cholesteryl esters17 triacylglycerol2 Synonyms: Bile salt activated lipase, sterol esterase, 4-nitrophenyl butyrate7 triolein9 carboxyl ester lipase, steryl-ester acylhydrolase 2-(diethylamino)-4H-3,1-benzoxazin-4-one18 2-(diethylamino)-4H-thieno[2,3-d]oxazin-4-one18 Product Description 7 Excess cholesterol is stored intracellularly as KM: 4-nitrophenyl butyrate cholesterol esters. Cholesterol esterase (CE) is a 0.37 mM (with taurocholate) reversible enzyme that can hydrolyze or synthesize 0.73 mM (without taurocholate) fatty acid esters of cholesterol and other sterols. Hydrolysis of water insoluble long chain fatty acid Activators: esters requires bile salt activation. Hydrolysis of water ethanol9 methanol9 soluble esters of short chain fatty acids and n-butanol9 sodium taurocholate10 lysophospholipids does not require activation by bile sodium cholate11 salts.1 Cholesterol esterase catalyzes the following reaction: Inhibitors: bisphenol A methacrylate12 Hg2+,15 Cholesterol Cholesterol Cholesterol diisopropylfluorophosphate13 enolase14 esters esterase + Fatty acid sodium fluoride15 phosphatidic acid16 phosphatidylcholine16 phosphatidylserine16 While found primarily in the pancreas and pancreatic 4-nitrophenyl-N-substituted carbamates11 fluid, it occurs in other tissues as well. In the bovine adrenal cortex, this reaction is one of the rate limiting Ki: steps in steroidogenesis, involving the release of 4-nitrophenyl-N-allyl -
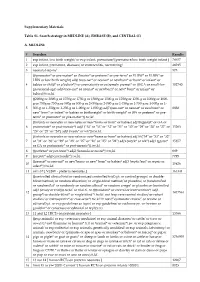
EMBASE (B), and CENTRAL (C) A. MEDLINE # Searches
Supplementary Materials Table S1. Search strategy in MEDLINE (A), EMBASE (B), and CENTRAL (C) A. MEDLINE # Searches Results 1 exp infant, low birth weight/ or exp infant, premature/ [premature/low birth weight infant ] 78657 2 exp infant, premature, diseases/ or enterocolitis, necrotizing/ 46015 3 neonatal sepsis/ 575 (((prematur* or pre-matur* or i?matur* or preterm* or pre-term* or VLBW* or ELBW* or LBW or low birth weight) adj6 (neo-nat* or neonat* or newborn* or born* or infant* or 4 babies or child* or p?ediatr*)) or prematurity or extremely premat* or ((SGA or small-for- 102740 gestational-age) adj6 (neo-nat* or neonat* or newborn* or new* born* or infant* or babies))).tw,kf. ((2000g or 2000-g or 1750g or 1750-g or 1500g or 1500-g or 1250g or 1250-g or 1000g or 1000- g or 750g or 750-g or 500g or 500-g or 2-000g or 2-000-g or 1-750g or 1-750-g or 1-500g or 1- 5 500-g or 1-250g or 1-250-g or 1-000g or 1-000-g) adj7 (neo-nat* or neonat* or newborn* or 8838 new* born* or infant* or babies or birthweight* or birth weight* or BW or preterm* or pre- term* or prematur* or pre-matur*)).tw,kf. ((infants or neonates or neo-nates or new*borns or born* or babies) adj18 (gestat* or GA or 6 postmenstr* or post-menstr*) adj3 ("34" or "33" or "32" or "31" or "30" or "29" or "28" or "27" or 15263 "26" or "25" or "24") adj3 (week* or wk*)).tw,kf. -

Cholesterol Esterase from Pseudomonas Sp
Cholesterol Esterase from Pseudomonas sp. Catalog Number C9281 Storage Temperature –20 C CAS RN 9026-00-0 Specific activity: 10,000 units/g protein EC 3.1.1.13 Synonyms: Bile salt activated lipase, sterol esterase, Unit definition: one unit will hydrolyze 1.0 mole of carboxyl ester lipase, steryl-ester acylhydrolase cholesteryl oleate to cholesterol and oleic acid per minute at pH 7.0 at 37 C in the presence of Product Description taurocholate. Excess cholesterol is stored intracellularly as cholesterol esters. Cholesterol esterase (CE) is a Cholesterol esterase is assayed spectrophotometrically reversible enzyme, with a molecular mass of in a 3.0 mL reaction mixture containing 287 mM 1 129 kDa, that can hydrolyze or synthesize fatty acid potassium phosphate, pH 7.0, 0.25% (w/v) taurocholic esters of cholesterol and other sterols. Hydrolysis of acid, 0.25% (w/v) cholic acid, 4-6 units peroxidase, water-insoluble long chain fatty acid esters requires bile 1.4 mM cholesteryl oleate, 1.7% (v/v) polyoxyethylene salt activation. Hydrolysis of water-soluble esters of 9-lauryl ether, 0.14% (w/v) NaCl, 0.083% (w/v) phenol, short chain fatty acids and lysophospholipids does not 0.03% (w/v) 4-aminoantipyrine, 1-1.5 units cholesterol 2 require activation by bile salts. Cholesterol esterase oxidase, and 0.013-0.143 unit cholesterol esterase. catalyzes the following reaction: Precautions and Disclaimer Cholesterol Cholesterol Cholesterol This product is for R&D use only, not for drug, esters esterase + Fatty acid household, or other uses. Please consult the Safety Data Sheet for information regarding hazards and safe In the bovine adrenal cortex, this reaction is one of the handling practices. -

Disease Models & Mechanisms • DMM • Accepted Manuscript
Transcriptomic analyses of gastrulation-stage C57BL/6J and C57BL/6NHsd mouse embryos with differential susceptibility to alcohol Karen E. Boschen1, Travis S. Ptacek2,3, Matthew E. Berginski4, Jeremy M. Simon2,3,5, Scott E. Parnell1,6,7* 1Bowles Center for Alcohol Studies, The University of North Carolina at Chapel Hill, Chapel Hill, NC, USA 2Carolina Institute for Developmental Disabilities, The University of North Carolina at Chapel Hill, Chapel Hill, NC, USA 3UNC Neuroscience Center, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA 4 Department of Pharmacology, The University of North Carolina at Chapel Hill, Chapel Hill, NC, USA 5 Department of Genetics, The University of North Carolina at Chapel Hill, Chapel Hill, NC, USA 6 Department of Cell Biology and Physiology, The University of North Carolina at Chapel Hill, Chapel Hill, NC, USA 7 Carolina Institute for Developmental Disabilities, The University of North Carolina at Chapel Hill, Chapel Hill, NC, USA *Address correspondence to: Scott E. Parnell, Ph.D. Bowles Center for Alcohol Studies University of North Carolina 104 Manning Drive CB# 7178 Thurston Bowles Chapel Hill, NC 27599 Phone: 919-966-8195, Fax: 919-966-5679 Email: [email protected], ORCID: 0000-0003-2038-6548 © 2021. Published by The Company of Biologists Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution and reproduction in any medium provided that the original work is properly attributed. Disease Models & Mechanisms • DMM Accepted manuscript SUMMARY STATEMENT RNA-seq in gastrulation-stage mouse embryos provides information about gene expression patterns during normal mouse development and evidence that pre-existing genetic variability mediates risk to prenatal alcohol-induced birth defects. -

Characterization and Application of a Sterol Esterase Immobilized on Polyacrylate Epoxy-Activated Carriers
* Manuscript Characterization and application of a sterol esterase immobilized on polyacrylate epoxy-activated carriers (DilbeadsTM) P. Torresa, A. Datlab, V.W. Rajasekarb, S. Zambreb, T. Asharb, M. Yatesa, M.L. Rojas-Cervantesc, O. Calero-Ruedad, V. Barbad, M.J. Martínezd, A. Ballesterosa and F.J. Ploua,* a Instituto de Catálisis y Petroleoquímica, CSIC, Cantoblanco, 28049 Madrid, Spain. b Research Support International Ltd., DILComplex, Thane west, India. c Departamento de Química Inorgánica y Química Técnica, Facultad de Ciencias, UNED, 28040 Madrid, Spain. d Centro de Investigaciones Biológicas, CSIC, Ramiro de Maeztu 9, 28040 Madrid, Spain. * Correspondence author: Francisco J. Plou, Departamento de Biocatálisis, Instituto de Catálisis y Petroleoquímica, CSIC, Cantoblanco, Marie Curie 2, 28049 Madrid, Spain. Phone: +34-91-5854869; Fax: +34-91-5854760. E-mail: [email protected]. URL: http://www.icp.csic.es/abg 1 ABSTRACT The sterol esterase from the ascomycete Ophiostoma piceae was immobilized on novel polyacrylate-based epoxy-activated carriers (DilbeadsTM). Six supports with particle sizes between 120-165 m were prepared varying the composition of monomers, crosslinkers and porogens. Their surface areas and porosities were determined by N2 adsorption and mercury intrusion porosimetry. The pore volumes ranged from 0.63 to 1.32 cm3/g, but only DilbeadsTM RS and NK had narrow pore size distributions (with maxima at 33.5 and 67.0 nm, respectively). The distribution of the enzyme in the support was studied by fluorescence confocal microscopy. The immobilized esterase on DilbeadsTM TA showed a significant pH and thermal stability and was assayed in the continuous hydrolysis of cholesteryl esters -present in the pulp industry process waters-. -

WO 2016/206838 Al 29 December 2016 (29.12.2016) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2016/206838 Al 29 December 2016 (29.12.2016) P O P C T (51) International Patent Classification: Unilever R&D Port Sunlight Quarry Road East, Bebington, CUD 1/83 (2006.01) CUD 11/00 (2006.01) Wirral Merseyside CH63 3JW (GB). CUD 3/37 (2006.01) C D 1/72 (2006.01) (74) Agent: AVILA, David, Victor; Unilever Patent Group CUD 3/386 (2006.01) Colworth House, Sharnbrook, Bedford, Bedfordshire (21) International Application Number: MK44 1LQ (GB). PCT/EP2016/059425 (81) Designated States (unless otherwise indicated, for every (22) International Filing Date: kind of national protection available): AE, AG, AL, AM, 27 April 2016 (27.04.2016) AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, (25) Filing Language: English DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, (26) Publication Language: English HN, HR, HU, ID, IL, IN, IR, IS, JP, KE, KG, KN, KP, KR, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, MG, (30) Priority Data: MK, MN, MW, MX, MY, MZ, NA, NG, NL NO, NZ, OM, 15 174015.6 26 June 2015 (26.06.2015) EP PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, SC, (71) Applicant (for AE, AG, AU, BB, BH, BN, BW, BZ, CA, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, CY, EG, GB, GD, GH, GM, IE, IL, IN, KE, KN, LC, LK, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. -

Gfapind Nestin G FA P DA PI
Figure S1 GFAPInd NB +bFGF/+EGF NB -bFGF/-EGF nestin GFAP DAPI (a) Figure S1 GFAPConst NB +bFGF+/+EGF NB -bFGF/-EGF nestin GFAP DAPI (b) Figure S2 +bFGF/+EGF (a) Figure S2 -bFGF/-EGF (b) Figure S3 #10 #1095 #1051 #1063 #1043 #1083 ~20 weeks GSCs GSC_IRs compare tumor-propagating capacity proliferation gene expression Supplemental Table S1. Expression patterns of nestin and GFAP in GSCs self-renewing in vitro. GSC line nestin (%) GFAP (%) #10 90 ± 1,9 26 ± 5,8 #1095 92 ± 5,4 1,45 ± 0,1 #1063 74,4 ± 27,9 3,14 ± 1,8 #1051 92 ± 1,3 < 1 #1043 96,7 ± 1,4 96,7 ± 1,4 #1080 99 ± 0,6 99 ± 0,6 #1083 64,5 ± 14,1 64,5 ± 14,1 G112-NB 99 ± 1,6 99 ± 1,6 Immunofluorescence staining of GSCs cultured under self-renewal promoting condition Supplemental Table S2. Gene expression analysis by Gene Ontology terms. -

Supplementary Tables.Xlsx
Supplementary table S4 – Comparison of the ScHaa1- and CgHaa1- dependent regulatory networks. The dataset of C. glabrata genes herein found to be regulated by CgHaa1 under acetic acid stress was compared with the dataset of genes described to be regulated by ScHaa1, based on the information available at the YEASTRACT database. Identification of orthologues was performed using the Yeastmine algorithm available at the Candida Genome database website after which a manual curation of the data based on BLASTP was performed. The promoter region of S. cerevisiae or C. glabrata genes was searched for the described ScHaa1 binding site (5’-(G/C)(A/C)GG(G/C)-3’, designated Haa1-responsive element – HRE; Mira et al., 2011) using the DNA pattern matching algorithms embedded in the RSA tools website. Cells highlighted in grey correspond to the S. cerevisiae genes found to be regulated by ScHaa1 specifically under acetic acid stress, based on the dataset of Mira et al., (2010). Acetic acid-responsive genes regulated by ScHaa1 only C. glabrata S. cerevisiae S. cerevisiae ORF Function promoter harbors promoter harbors orthologue an HRE motif an HRE motif Ortholog(s) have actin binding, cytoskeletal regulatory protein binding, enzyme activator CAGL0A00891g YLR319C No No activity Ortholog(s) have role in double-strand break repair, meiotic sister chromatid segregation, CAGL0A00913g recombinational repair, replication fork processing and Cul8-RING ubiquitin ligase complex, YLR320W No No nucleus localization CAGL0A01001g No Description Available YLR326W Yes -
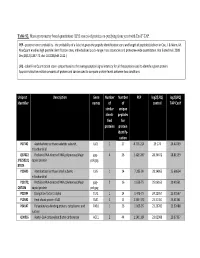
Table S2. Mass Spectrometry Based Quantitation (LFQ Scores) of Proteins Co-Purifying from Yeast with Exoy-TAP
Table S2. Mass spectrometry based quantitation (LFQ scores) of proteins co-purifying from yeast with ExoY-TAP. PEP - posterior error probability - the probability of a false hit given the peptide identification score and length of peptides (defined in Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide quantitation. Nat Biotechnol. 2008 Dec;26(12):1367-72. doi: 10.1038/nbt.1511.) LFQ - Label Free Quantitation score - proportional to the average peptide signal intensity for all the peptides used to identify a given protein. Approximates the relative amounts of protein and can be used to compare protein levels between two conditions. Uniprot Description Gene Number Number PEP log2(LFQ) log2(LFQ) identifier names of of control TAP-ExoY similar unique identi- peptides fied for proteins protein identify- cation P07342 Acetolactate synthase catalytic subunit, ILV2 1 27 8.77E-214 29.174 26.40759 mitochondrial Q87022 Probable RNA-directed RNA polymerase;Major gag- 4 26 1.42E-267 28.34672 28.81559 ;P32503;Q capsid protein pol;gag 87024 P25605 Acetolactate synthase small subunit, ILV6 1 14 7.28E-99 28.14865 25.68654 mitochondrial P23172; Probable RNA-directed RNA polymerase;Major gag- 216 1.63E-75 25.06163 20.40561 Q87026 capsid protein pol;gag P02994 Elongation factor 1-alpha TEF1 1 14 1.47E-75 24.22957 25.30567 P10591 Heat shock protein SSA1 SSA1 2 8 2.54E-173 23.31451 25.41361 P04147 Polyadenylate-binding protein, cytoplasmic and PAB1 126 1.06E-95 23.29782 23.92488 nuclear Q00955 Acetyl-CoA carboxylase;Biotin carboxylase ACC1 2 44 1.54E-164 23.02989 20.97337 P0C2I6;P0 Transposon Ty1-LR3 Gag-Pol polyprotein;Capsid TY1B- 57 3 1.88E-88 22.48395 23.64713 C2I9;P0C2 protein;Ty1 protease;Integrase; Reverse LR3;TY1B- J1;P47098 transcriptase/ribonuclease H (Rev. -
Generate Metabolic Map Poster
Authors: J. Michael Cherry Eurie Hong Benjamin Vincent Cynthia Krieger An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Rama Balakrishnan Ron Caspi Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. YeastCyc: Saccharomyces cerevisiae S288c Cellular Overview Connections between pathways are omitted for legibility.