Lakhtin V., Lakhtin M., Aleshkin V. INTERACTION of ESTERASES
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

MIAMI UNIVERSITY the Graduate School Certificate for Approving The
MIAMI UNIVERSITY The Graduate School Certificate for Approving the Dissertation We hereby approve the Dissertation of Matthew F. Rouhier Candidate for the Degree: Doctor of Philosophy ___________________________________________________________ Director (Dr. Ann Hagerman) ___________________________________________________________ Reader (Dr.Chris Makaroff) ___________________________________________________________ Reader (Dr.Gary Lorigan) __________________________________________________________ Reader (Dr. Richard Taylor) _________________________________________________________ Graduate School Representative (Dr. Paul James) ! ABSTRACT CHARACTERIZATION OF YDR036C FROM Saccharomyces cerevisiae by Matthew F. Rouhier Beta-hydroxyisobutyryl-CoA (HIBYL-CoA) hydrolases are found ubiquitously in eukaryotes where they function in the catabolism of valine. A homologous enzyme (YDR036C) is also present in yeast where valine catabolism is distinctly different and does not require a HIBYL-CoA hydrolase. Like the other eukaryotic hydrolases, the yeast hydrolase is a member of the crotonase super-family which catalyzes various reactions using CoA thioester substrates. Crotonases typically function in fatty acid degradation which is peroxisomal in yeast while YDR036C is found strictly in mitochondria. Like other HIBYL-CoA hydrolases the yeast enzyme demonstrated activity toward beta-hydroxyacyl-CoAs, but unlike the other HIBYL-CoA hydrolases the yeast activity is greatest with beta-hydroxypropionyl-CoA (3-HP-CoA). Further characterization of the active site has determined that the preference of the hydrolases for 3-HP- CoA or HIBYL-CoA is dependent upon the residue at position 177. The glutamate at position 121 is responsible for the coordination with the beta-hydroxyl group and replacement with valine renders the enzyme non-specific for a hydroxyl group. Another unique feature to yeast hydrolases is the presence of a C-terminal tail of 80 amino acids, which when removed renders the enzyme inactive, suggesting that it may play a role in substrate binding. -

Carcinogen-Induced Early Molecular Events and Its Implication In
Biochimica et Biophysica Acta 1772 (2007) 48–59 www.elsevier.com/locate/bbadis Carcinogen-induced early molecular events and its implication in the initiation of chemical hepatocarcinogenesis in rats: Chemopreventive role of vanadium on this process Tridib Chakraborty a, Amrita Chatterjee a, Ajay Rana b, Duraisami Dhachinamoorthi a, ⁎ Ashok Kumar P a, Malay Chatterjee a, a Division of Biochemistry, Department of Pharmaceutical Technology, Jadavpur University, PO Box 17028, Calcutta-700032, India b Division of Molecular Cardiology, Cardiovascular Research Institute, College of Medicine, The Texas A&M University System HSC, Temple, TX 76504, USA Received 21 May 2006; received in revised form 19 September 2006; accepted 16 October 2006 Available online 10 November 2006 Abstract Carcinogen-induced formation of DNA adducts and other types of DNA lesions are the critical molecular events in the initiation of chemical carcinogenesis and modulation of such events by chemopreventive agents could be an important step in limiting neoplastic transformation in vivo. Vanadium, a dietary micronutrient has been found to be effective in several types of cancers both in vivo and in vitro and also possesses profound anticarcinogenicity against rat models of mammary, colon and hepatocarcinogenesis. Presently, we report the chemopreventive potential of vanadium on diethylnitrosamine (DEN)-induced early DNA damages in rat liver. Hepatocarcinogenesis was induced in male Sprague–Dawley rats with a single, necrogenic, intraperitoneal (i.p.) injection of DEN (200 mg/kg body weight) at week 4. There was a significant induction of tissue- specific ethylguanines, steady elevation of modified DNA bases 8-hydroxy-2′-deoxyguanosines (8-OHdGs) (P<0.0001; 89.93%) along with substantial increment of the extent of single-strand breaks (SSBs) (P<0.0001) following DEN exposure. -

(10) Patent No.: US 8119385 B2
US008119385B2 (12) United States Patent (10) Patent No.: US 8,119,385 B2 Mathur et al. (45) Date of Patent: Feb. 21, 2012 (54) NUCLEICACIDS AND PROTEINS AND (52) U.S. Cl. ........................................ 435/212:530/350 METHODS FOR MAKING AND USING THEMI (58) Field of Classification Search ........................ None (75) Inventors: Eric J. Mathur, San Diego, CA (US); See application file for complete search history. Cathy Chang, San Diego, CA (US) (56) References Cited (73) Assignee: BP Corporation North America Inc., Houston, TX (US) OTHER PUBLICATIONS c Mount, Bioinformatics, Cold Spring Harbor Press, Cold Spring Har (*) Notice: Subject to any disclaimer, the term of this bor New York, 2001, pp. 382-393.* patent is extended or adjusted under 35 Spencer et al., “Whole-Genome Sequence Variation among Multiple U.S.C. 154(b) by 689 days. Isolates of Pseudomonas aeruginosa” J. Bacteriol. (2003) 185: 1316 1325. (21) Appl. No.: 11/817,403 Database Sequence GenBank Accession No. BZ569932 Dec. 17. 1-1. 2002. (22) PCT Fled: Mar. 3, 2006 Omiecinski et al., “Epoxide Hydrolase-Polymorphism and role in (86). PCT No.: PCT/US2OO6/OOT642 toxicology” Toxicol. Lett. (2000) 1.12: 365-370. S371 (c)(1), * cited by examiner (2), (4) Date: May 7, 2008 Primary Examiner — James Martinell (87) PCT Pub. No.: WO2006/096527 (74) Attorney, Agent, or Firm — Kalim S. Fuzail PCT Pub. Date: Sep. 14, 2006 (57) ABSTRACT (65) Prior Publication Data The invention provides polypeptides, including enzymes, structural proteins and binding proteins, polynucleotides US 201O/OO11456A1 Jan. 14, 2010 encoding these polypeptides, and methods of making and using these polynucleotides and polypeptides. -

(12) Patent Application Publication (10) Pub. No.: US 2012/0058468 A1 Mickeown (43) Pub
US 20120058468A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2012/0058468 A1 MickeoWn (43) Pub. Date: Mar. 8, 2012 (54) ADAPTORS FOR NUCLECACID Related U.S. Application Data CONSTRUCTS IN TRANSMEMBRANE (60) Provisional application No. 61/148.737, filed on Jan. SEQUENCING 30, 2009. Publication Classification (75) Inventor: Brian Mckeown, Oxon (GB) (51) Int. Cl. CI2O I/68 (2006.01) (73) Assignee: OXFORD NANOPORE C7H 2L/00 (2006.01) TECHNOLGIES LIMITED, (52) U.S. Cl. ......................................... 435/6.1:536/23.1 Oxford (GB) (57) ABSTRACT (21) Appl. No.: 13/147,159 The invention relates to adaptors for sequencing nucleic acids. The adaptors may be used to generate single stranded constructs of nucleic acid for sequencing purposes. Such (22) PCT Fled: Jan. 29, 2010 constructs may contain both strands from a double stranded deoxyribonucleic acid (DNA) or ribonucleic acid (RNA) (86) PCT NO.: PCT/GB1O/OO160 template. The invention also relates to the constructs gener ated using the adaptors, methods of making the adaptors and S371 (c)(1), constructs, as well as methods of sequencing double stranded (2), (4) Date: Nov. 15, 2011 nucleic acids. Patent Application Publication Mar. 8, 2012 Sheet 1 of 4 US 2012/0058468 A1 Figure 5' 3 Figure 2 Figare 3 Patent Application Publication Mar. 8, 2012 Sheet 2 of 4 US 2012/0058468 A1 Figure 4 End repair Acid adapters ligate adapters Patent Application Publication Mar. 8, 2012 Sheet 3 of 4 US 2012/0058468 A1 Figure 5 Wash away type ifype foducts irrirrosilise Type| capture WashRE products away unbcure Pace É: Wash away unbound rag terts Free told fasgirre?ts aid raiser to festible Figure 6 Beatre Patent Application Publication Mar. -

Cholesterol Esterase from Porcine Pancreas
Cholesterol Esterase from porcine pancreas Catalog Number C9464 Storage Temperature –20 °C CAS RN 9026-00-0 Substrates: EC 3.1.1.13 cholesteryl esters17 triacylglycerol2 Synonyms: Bile salt activated lipase, sterol esterase, 4-nitrophenyl butyrate7 triolein9 carboxyl ester lipase, steryl-ester acylhydrolase 2-(diethylamino)-4H-3,1-benzoxazin-4-one18 2-(diethylamino)-4H-thieno[2,3-d]oxazin-4-one18 Product Description 7 Excess cholesterol is stored intracellularly as KM: 4-nitrophenyl butyrate cholesterol esters. Cholesterol esterase (CE) is a 0.37 mM (with taurocholate) reversible enzyme that can hydrolyze or synthesize 0.73 mM (without taurocholate) fatty acid esters of cholesterol and other sterols. Hydrolysis of water insoluble long chain fatty acid Activators: esters requires bile salt activation. Hydrolysis of water ethanol9 methanol9 soluble esters of short chain fatty acids and n-butanol9 sodium taurocholate10 lysophospholipids does not require activation by bile sodium cholate11 salts.1 Cholesterol esterase catalyzes the following reaction: Inhibitors: bisphenol A methacrylate12 Hg2+,15 Cholesterol Cholesterol Cholesterol diisopropylfluorophosphate13 enolase14 esters esterase + Fatty acid sodium fluoride15 phosphatidic acid16 phosphatidylcholine16 phosphatidylserine16 While found primarily in the pancreas and pancreatic 4-nitrophenyl-N-substituted carbamates11 fluid, it occurs in other tissues as well. In the bovine adrenal cortex, this reaction is one of the rate limiting Ki: steps in steroidogenesis, involving the release of 4-nitrophenyl-N-allyl -

(5'-Uridylyl)Tyrosine Is the Bond Between the Genome-Linked Protein and the RNA of Poliovirus
Proc. Natl. Acad. Sci. USA Vol. 75, No 10, pp. 4868-4872, October 1978 Biochemistry 04-(5'-Uridylyl)tyrosine is the bond between the genome-linked protein and the RNA of poliovirus* (iodination/acid and alkali hydrolysis/enzymatic degradation/paper electrophoresis and chromatography) PAUL G. ROTHBERG, TIMOTHY J. R. HARRISt, AKIO NOMOTOt, AND ECKARD WIMMER Department of Microbiology, School of Basic Health Sciences, State University of New York at Stony Brook, Stony Brook, Long Island, New York 11794 Communicated by Seymour S. Cohen, August 3, 1978 ABSTRACT Virion RNA of poliovirus type 1 has been an- MATERIALS AND METHODS alyzed for the linkage between genome-protein VPg and the polyribonucleotide chain. Hydrolysis of the linkage with acid Poliovirus was grown and labeled with phosphorus-32 in HeLa or alkali and enzymatic degradation lead to the conclusion that cell suspension cultures as previously described (9). VPg was the bond is neither a phosphodiester such as nucleotidyl-(P- labeled with [3H]tyrosine as follows: 2.5 X 109 HeLa cells were O-serine (or threonine) nor a phosphoramidate such as washed twice with Earle's saline, suspended in 200 ml of Earle's nucleotidyl4(P-N)amino acid. VPg-RNA can be iodinated by the saline containing 1 mg of actinomycin D, and infected with 50 Bolton and Hunter reagent liodinated 3(4-hydroxyphenyl)pro- plaque-forming units of PV1 per cell. After 25 min at, room pionic acid N-hydroxysuccinimide ester] but not by the chlo- temperature, 200 ml of medium containing 168 ml of Earle's ramine-T or lactoperoxidase procedures, an observation saline, 4 ml of an antibiotic solution (10,000 units of penicillin suggesting that VPg does not contain accessible tyrosine. -

Proquest Dissertations
Characterizing the Endoribonuclease Activity of APE1 Wan Cheol Kim BSc, Simon Fraser University, 2007 Thesis Submitted in Partial Fulfillment of The Requirements for the Degree of Master of Science In Mathematical, Computer, and Physical Sciences (Chemistry) The University of Northern British Columbia June 2009 © Wan Cheol Kim, 2009 Library and Archives Bibliotheque et 1*1 Canada Archives Canada Published Heritage Direction du Branch Patrimoine de I'edition 395 Wellington Street 395, rue Wellington OttawaONK1A0N4 Ottawa ON K1A 0N4 Canada Canada Your file Votre reference ISBN: 978-0-494-60814-2 Our file Notre reference ISBN: 978-0-494-60814-2 NOTICE: AVIS: The author has granted a non L'auteur a accorde une licence non exclusive exclusive license allowing Library and permettant a la Bibliotheque et Archives Archives Canada to reproduce, Canada de reproduire, publier, archiver, publish, archive, preserve, conserve, sauvegarder, conserver, transmettre au public communicate to the public by par telecommunication ou par I'lnternet, preter, telecommunication or on the Internet, distribuer et vendre des theses partout dans le loan, distribute and sell theses monde, a des fins commerciales ou autres, sur worldwide, for commercial or non support microforme, papier, electronique et/ou commercial purposes, in microform, autres formats. paper, electronic and/or any other formats. The author retains copyright L'auteur conserve la propriete du droit d'auteur ownership and moral rights in this et des droits moraux qui protege cette these. Ni thesis. Neither the thesis nor la these ni des extraits substantiels de celle-ci substantial extracts from it may be ne doivent etre imprimes ou autrement printed or otherwise reproduced reproduits sans son autorisation. -
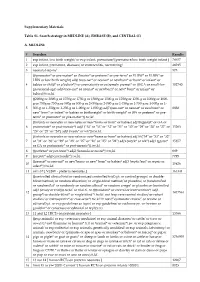
EMBASE (B), and CENTRAL (C) A. MEDLINE # Searches
Supplementary Materials Table S1. Search strategy in MEDLINE (A), EMBASE (B), and CENTRAL (C) A. MEDLINE # Searches Results 1 exp infant, low birth weight/ or exp infant, premature/ [premature/low birth weight infant ] 78657 2 exp infant, premature, diseases/ or enterocolitis, necrotizing/ 46015 3 neonatal sepsis/ 575 (((prematur* or pre-matur* or i?matur* or preterm* or pre-term* or VLBW* or ELBW* or LBW or low birth weight) adj6 (neo-nat* or neonat* or newborn* or born* or infant* or 4 babies or child* or p?ediatr*)) or prematurity or extremely premat* or ((SGA or small-for- 102740 gestational-age) adj6 (neo-nat* or neonat* or newborn* or new* born* or infant* or babies))).tw,kf. ((2000g or 2000-g or 1750g or 1750-g or 1500g or 1500-g or 1250g or 1250-g or 1000g or 1000- g or 750g or 750-g or 500g or 500-g or 2-000g or 2-000-g or 1-750g or 1-750-g or 1-500g or 1- 5 500-g or 1-250g or 1-250-g or 1-000g or 1-000-g) adj7 (neo-nat* or neonat* or newborn* or 8838 new* born* or infant* or babies or birthweight* or birth weight* or BW or preterm* or pre- term* or prematur* or pre-matur*)).tw,kf. ((infants or neonates or neo-nates or new*borns or born* or babies) adj18 (gestat* or GA or 6 postmenstr* or post-menstr*) adj3 ("34" or "33" or "32" or "31" or "30" or "29" or "28" or "27" or 15263 "26" or "25" or "24") adj3 (week* or wk*)).tw,kf. -

Manual D'estil Per a Les Ciències De Laboratori Clínic
MANUAL D’ESTIL PER A LES CIÈNCIES DE LABORATORI CLÍNIC Segona edició Preparada per: XAVIER FUENTES I ARDERIU JAUME MIRÓ I BALAGUÉ JOAN NICOLAU I COSTA Barcelona, 14 d’octubre de 2011 1 Índex Pròleg Introducció 1 Criteris generals de redacció 1.1 Llenguatge no discriminatori per raó de sexe 1.2 Llenguatge no discriminatori per raó de titulació o d’àmbit professional 1.3 Llenguatge no discriminatori per raó d'ètnia 2 Criteris gramaticals 2.1 Criteris sintàctics 2.1.1 Les conjuncions 2.2 Criteris morfològics 2.2.1 Els articles 2.2.2 Els pronoms 2.2.3 Els noms comuns 2.2.4 Els noms propis 2.2.4.1 Els antropònims 2.2.4.2 Els noms de les espècies biològiques 2.2.4.3 Els topònims 2.2.4.4 Les marques registrades i els noms comercials 2.2.5 Els adjectius 2.2.6 El nombre 2.2.7 El gènere 2.2.8 Els verbs 2.2.8.1 Les formes perifràstiques 2.2.8.2 L’ús dels infinitius ser i ésser 2.2.8.3 Els verbs fer, realitzar i efectuar 2.2.8.4 Les formes i l’ús del gerundi 2.2.8.5 L'ús del verb haver 2.2.8.6 Els verbs haver i caldre 2.2.8.7 La forma es i se davant dels verbs 2.2.9 Els adverbis 2.2.10 Les locucions 2.2.11 Les preposicions 2.2.12 Els prefixos 2.2.13 Els sufixos 2.2.14 Els signes de puntuació i altres signes ortogràfics auxiliars 2.2.14.1 La coma 2.2.14.2 El punt i coma 2.2.14.3 El punt 2.2.14.4 Els dos punts 2.2.14.5 Els punts suspensius 2.2.14.6 El guionet 2.2.14.7 El guió 2.2.14.8 El punt i guió 2.2.14.9 L’apòstrof 2.2.14.10 L’interrogant 2 2.2.14.11 L’exclamació 2.2.14.12 Les cometes 2.2.14.13 Els parèntesis 2.2.14.14 Els claudàtors 2.2.14.15 -

Cholesterol Esterase from Pseudomonas Sp
Cholesterol Esterase from Pseudomonas sp. Catalog Number C9281 Storage Temperature –20 C CAS RN 9026-00-0 Specific activity: 10,000 units/g protein EC 3.1.1.13 Synonyms: Bile salt activated lipase, sterol esterase, Unit definition: one unit will hydrolyze 1.0 mole of carboxyl ester lipase, steryl-ester acylhydrolase cholesteryl oleate to cholesterol and oleic acid per minute at pH 7.0 at 37 C in the presence of Product Description taurocholate. Excess cholesterol is stored intracellularly as cholesterol esters. Cholesterol esterase (CE) is a Cholesterol esterase is assayed spectrophotometrically reversible enzyme, with a molecular mass of in a 3.0 mL reaction mixture containing 287 mM 1 129 kDa, that can hydrolyze or synthesize fatty acid potassium phosphate, pH 7.0, 0.25% (w/v) taurocholic esters of cholesterol and other sterols. Hydrolysis of acid, 0.25% (w/v) cholic acid, 4-6 units peroxidase, water-insoluble long chain fatty acid esters requires bile 1.4 mM cholesteryl oleate, 1.7% (v/v) polyoxyethylene salt activation. Hydrolysis of water-soluble esters of 9-lauryl ether, 0.14% (w/v) NaCl, 0.083% (w/v) phenol, short chain fatty acids and lysophospholipids does not 0.03% (w/v) 4-aminoantipyrine, 1-1.5 units cholesterol 2 require activation by bile salts. Cholesterol esterase oxidase, and 0.013-0.143 unit cholesterol esterase. catalyzes the following reaction: Precautions and Disclaimer Cholesterol Cholesterol Cholesterol This product is for R&D use only, not for drug, esters esterase + Fatty acid household, or other uses. Please consult the Safety Data Sheet for information regarding hazards and safe In the bovine adrenal cortex, this reaction is one of the handling practices. -

Mimicking the Action of Ribonucleases: Studies on Rnase a and Design of PNA Based Artificial Enzymes
From DEPARTMENT OF BIOSCIENCES AND NUTRITION Karolinska Institutet, Stockholm, Sweden Mimicking the action of ribonucleases: studies on RNase A and design of PNA based artificial enzymes Alice Ghidini Stockholm 2015 All previously published papers were reproduced with permission from the publisher. Published by Karolinska Institutet. Printed by Eprint AB 2015 © Alice Ghidini, 2015 ISBN 978-91-7676-039-0 Mimicking the action of ribonucleases: studies on RNase A and design of PNA based artificial enzymes THESIS FOR DOCTORAL DEGREE (Ph.D.) By Alice Ghidini Principal Supervisor: Opponent: Professor Roger Strömberg Professor Michael J. Gait Karolinska Institutet Medical Research Council Department of Bioscience and nutrition Department of Laboratory of Molecular Biology Division of Bioorganic chemistry Examination Board: Co-supervisor(s): Lars Baltzer Merita Murtola Uppsala Universitet Karolinska Institutet Department of Institutionen för kemi Department of Bioscience and nutrition Division of BMC, Fysikalisk organisk kemi Division of Bioorganic chemistry Mikael Leijon Malgorzata Honcharenko National Veterinary Institute (SVA) Karolinska Institutet Division of Virology, Immunobiology and Department of Bioscience and nutrition Parasitology (VIP) Division of Bioorganic chemistry Marcus Wilhelmsson Chalmers University of Technology Department of Chemical and Chemical Engineering/Chemistry and Biochemistry With love to my family, lontani ma sempre vicini… ABSTRACT A 3’-deoxy-3’-C-methylenephosphonate modified diribonucleotide is highly resistant to degradation by spleen phosphodiesterase and not cleaved at all by snake venom phosphodiesterase. Despite that both the vicinal 2-hydroxy nucleophile and the 5’-oxyanion leaving group are intact, the 3’-methylenephosponate RNA modification is also highly resistant towards the action of RNase A. Several different approaches were explored for conjugation of oligoethers to PNA with internally or N-terminal placed diaminopropionic acid residues. -

Sequence Verification of Oligonucleotides Containing Multiple Arylamine Modifications by Enzymatic Digestion and Liquid Chromatography Mass Spectrometry (LC/MS)
Sequence Verification of Oligonucleotides Containing Multiple Arylamine Modifications by Enzymatic Digestion and Liquid Chromatography Mass Spectrometry (LC/MS) Lan Gao,a Li Zhang,b Bongsup P. Cho,b and M. Paul Chiarellia a Department of Chemistry, Loyola University, Chicago, Illinois, USA b Department of Biomedical and Pharmaceutical Sciences, College of Pharmacy, University of Rhode Island, Kingston, Rhode Island, USA An©analytical©method©for©the©structure©differentiation©of©arylamine©modified©oligonucleotides (ODNs)©using©on-line©LC/MS©analysis©of©raw©exonuclease©digests©is©described.©Six©different dodeca©ODNs©derived©from©the©reaction©of©N-acetoxy-N-(trifluoroacetyl)-2-aminofluorene with©the©dodeca©oligonucleotide©5=-CTCGGCGCCATC-3=©are©isolated©and©sequenced©with©this LC/MS©method©using©3=-©and©5=-exonucleases.©When©the©three©products©modified©by©a©single aminofluorene©(AF)©are©subjected©to©3=-exonuclease©digestion,©the©exonuclease©will©cleave©a modified©nucleotide©but©when©di-AF©modified©ODNs©are©analyzed©the©3=-exonuclease©ceases to©cleave©nucleotides©when©the©first©modification©is©exposed©at©the©3=-terminus.©Small abundances©of©ODN©fragments©formed©by©the©cleavage©of©an©AF-modified©nucleotide©were observed©when©two©of©the©three©di-AF©modified©ODNs©were©subjected©to©5=-exonuclease digestion.©The©results©of©the©5=-exonuclease©studies©of©the©three©di-AF©modified©ODNs©suggest that©as©the©number©of©unmodified©bases©between©two©modifications©in©an©ODN©sequence increases,©the©easier©it©becomes©to©sequence©beyond©the©modification©closest©to©the©5©=-terminus. The©results©of©this©study©indicate©that©the©LC/MS©method©described©here©would©be©useful©in sequencing©ODNs©modified©by©multiple©arylamines©to©be©used©as©templates©for©site-specific mutagenesis©studies.© (J©Am©Soc©Mass©Spectrom©2008,©19,©1147–1155)©©©2008©American©Society for©Mass©Spectrometry nterest©in©the©analysis©of©carcinogen-modified©nu- depend©on©the©number©of©guanine©bases©present©in©the cleic©acids©is©motivated©by©the©belief©that©the©forma- sequence.