Ecology and Physiology of Planktonic Archaea (Santoro)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Anoxygenic Photosynthesis in Photolithotrophic Sulfur Bacteria and Their Role in Detoxication of Hydrogen Sulfide
antioxidants Review Anoxygenic Photosynthesis in Photolithotrophic Sulfur Bacteria and Their Role in Detoxication of Hydrogen Sulfide Ivan Kushkevych 1,* , Veronika Bosáková 1,2 , Monika Vítˇezová 1 and Simon K.-M. R. Rittmann 3,* 1 Department of Experimental Biology, Faculty of Science, Masaryk University, 62500 Brno, Czech Republic; [email protected] (V.B.); [email protected] (M.V.) 2 Department of Biology, Faculty of Medicine, Masaryk University, 62500 Brno, Czech Republic 3 Archaea Physiology & Biotechnology Group, Department of Functional and Evolutionary Ecology, Universität Wien, 1090 Vienna, Austria * Correspondence: [email protected] (I.K.); [email protected] (S.K.-M.R.R.); Tel.: +420-549-495-315 (I.K.); +431-427-776-513 (S.K.-M.R.R.) Abstract: Hydrogen sulfide is a toxic compound that can affect various groups of water microorgan- isms. Photolithotrophic sulfur bacteria including Chromatiaceae and Chlorobiaceae are able to convert inorganic substrate (hydrogen sulfide and carbon dioxide) into organic matter deriving energy from photosynthesis. This process takes place in the absence of molecular oxygen and is referred to as anoxygenic photosynthesis, in which exogenous electron donors are needed. These donors may be reduced sulfur compounds such as hydrogen sulfide. This paper deals with the description of this metabolic process, representatives of the above-mentioned families, and discusses the possibility using anoxygenic phototrophic microorganisms for the detoxification of toxic hydrogen sulfide. Moreover, their general characteristics, morphology, metabolism, and taxonomy are described as Citation: Kushkevych, I.; Bosáková, well as the conditions for isolation and cultivation of these microorganisms will be presented. V.; Vítˇezová,M.; Rittmann, S.K.-M.R. -

Limits of Life on Earth Some Archaea and Bacteria
Limits of life on Earth Thermophiles Temperatures up to ~55C are common, but T > 55C is Some archaea and bacteria (extremophiles) can live in associated usually with geothermal features (hot springs, environments that we would consider inhospitable to volcanic activity etc) life (heat, cold, acidity, high pressure etc) Thermophiles are organisms that can successfully live Distinguish between growth and survival: many organisms can survive intervals of harsh conditions but could not at high temperatures live permanently in such conditions (e.g. seeds, spores) Best studied extremophiles: may be relevant to the Interest: origin of life. Very hot environments tolerable for life do not seem to exist elsewhere in the Solar System • analogs for extraterrestrial environments • `extreme’ conditions may have been more common on the early Earth - origin of life? • some unusual environments (e.g. subterranean) are very widespread Extraterrestrial Life: Spring 2008 Extraterrestrial Life: Spring 2008 Grand Prismatic Spring, Yellowstone National Park Hydrothermal vents: high pressure in the deep ocean allows liquid water Colors on the edge of the at T >> 100C spring are caused by different colonies of thermophilic Vents emit superheated water (300C or cyanobacteria and algae more) that is rich in minerals Hottest water is lifeless, but `cooler’ ~50 species of such thermophiles - mostly archae with some margins support array of thermophiles: cyanobacteria and anaerobic photosynthetic bacteria oxidize sulphur, manganese, grow on methane + carbon monoxide etc… Sulfolobus: optimum T ~ 80C, minimum 60C, maximum 90C, also prefer a moderately acidic pH. Live by oxidizing sulfur Known examples can grow (i.e. multiply) at temperatures which is abundant near hot springs. -
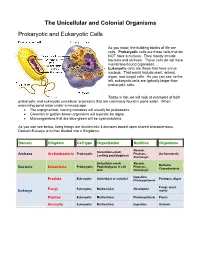
The Unicellular and Colonial Organisms Prokaryotic And
The Unicellular and Colonial Organisms Prokaryotic and Eukaryotic Cells As you know, the building blocks of life are cells. Prokaryotic cells are those cells that do NOT have a nucleus. They mostly include bacteria and archaea. These cells do not have membrane-bound organelles. Eukaryotic cells are those that have a true nucleus. That would include plant, animal, algae, and fungal cells. As you can see, to the left, eukaryotic cells are typically larger than prokaryotic cells. Today in lab, we will look at examples of both prokaryotic and eukaryotic unicellular organisms that are commonly found in pond water. When examining pond water under a microscope… The unpigmented, moving microbes will usually be protozoans. Greenish or golden-brown organisms will typically be algae. Microorganisms that are blue-green will be cyanobacteria. As you can see below, living things are divided into 3 domains based upon shared characteristics. Domain Eukarya is further divided into 4 Kingdoms. Domain Kingdom Cell type Organization Nutrition Organisms Absorb, Unicellular-small; Prokaryotic Photsyn., Archaeacteria Archaea Archaebacteria Lacking peptidoglycan Chemosyn. Unicellular-small; Absorb, Bacteria, Prokaryotic Peptidoglycan in cell Photsyn., Bacteria Eubacteria Cyanobacteria wall Chemosyn. Ingestion, Eukaryotic Unicellular or colonial Protozoa, Algae Protista Photosynthesis Fungi, yeast, Fungi Eukaryotic Multicellular Absorption Eukarya molds Plantae Eukaryotic Multicellular Photosynthesis Plants Animalia Eukaryotic Multicellular Ingestion Animals Prokaryotic Organisms – the archaea, non-photosynthetic bacteria, and cyanobacteria Archaea - Microorganisms that resemble bacteria, but are different from them in certain aspects. Archaea cell walls do not include the macromolecule peptidoglycan, which is always found in the cell walls of bacteria. Archaea usually live in extreme, often very hot or salty environments, such as hot mineral springs or deep-sea hydrothermal vents. -

Archaeal Distribution and Abundance in Water Masses of the Arctic Ocean, Pacific Sector
Vol. 69: 101–112, 2013 AQUATIC MICROBIAL ECOLOGY Published online April 30 doi: 10.3354/ame01624 Aquat Microb Ecol FREEREE ACCESSCCESS Archaeal distribution and abundance in water masses of the Arctic Ocean, Pacific sector Chie Amano-Sato1, Shohei Akiyama1, Masao Uchida2, Koji Shimada3, Motoo Utsumi1,* 1University of Tsukuba, Tennodai, Tsukuba, Ibaraki 305-8572, Japan 2National Institute for Environmental Studies, Onogawa, Tsukuba, Ibaraki 305-8506, Japan 3Tokyo University of Marine Science and Technology, Konan, Minato-ku, Tokyo 108-8477, Japan ABSTRACT: Marine planktonic Archaea have been recently recognized as an ecologically impor- tant component of marine prokaryotic biomass in the world’s oceans. Their abundance and meta- bolism are closely connected with marine geochemical cycling. We evaluated the distribution of planktonic Archaea in the Pacific sector of the Arctic Ocean using fluorescence in situ hybridiza- tion (FISH) with catalyzed reporter deposition (CARD-FISH) and performed statistical analyses using data for archaeal abundance and geochemical variables. The relative abundance of Thaum - archaeota generally increased with depth, and euryarchaeal abundance was the lowest of all planktonic prokaryotes. Multiple regression analysis showed that the thaumarchaeal relative abundance was negatively correlated with ammonium and dissolved oxygen concentrations and chlorophyll fluorescence. Canonical correspondence analysis showed that archaeal distributions differed with oceanographic water masses; in particular, Thaumarchaeota were abundant from the halocline layer to deep water, where salinity was higher and most nutrients were depleted. However, at several stations on the East Siberian Sea side of the study area and along the North- wind Ridge, Thaumarchaeota and Bacteria were proportionally very abundant at the bottom in association with higher nutrient conditions. -

Oceans of Archaea Abundant Oceanic Crenarchaeota Appear to Derive from Thermophilic Ancestors That Invaded Low-Temperature Marine Environments
Oceans of Archaea Abundant oceanic Crenarchaeota appear to derive from thermophilic ancestors that invaded low-temperature marine environments Edward F. DeLong arth’s microbiota is remarkably per- karyotes), Archaea, and Bacteria. Although al- vasive, thriving at extremely high ternative taxonomic schemes have been recently temperature, low and high pH, high proposed, whole-genome and other analyses E salinity, and low water availability. tend to support Woese’s three-domain concept. One lineage of microbial life in par- Well-known and cultivated archaea generally ticular, the Archaea, is especially adept at ex- fall into several major phenotypic groupings: ploiting environmental extremes. Despite their these include extreme halophiles, methanogens, success in these challenging habitats, the Ar- and extreme thermophiles and thermoacido- chaea may now also be viewed as a philes. Early on, extremely halo- cosmopolitan lot. These microbes philic archaea (haloarchaea) were exist in a wide variety of terres- first noticed as bright-red colonies trial, freshwater, and marine habi- Archaea exist in growing on salted fish or hides. tats, sometimes in very high abun- a wide variety For many years, halophilic isolates dance. The oceanic Marine Group of terrestrial, from salterns, salt deposits, and I Crenarchaeota, for example, ri- freshwater, and landlocked seas provided excellent val total bacterial biomass in wa- marine habitats, model systems for studying adap- ters below 100 m. These wide- tations to high salinity. It was only spread Archaea appear to derive sometimes in much later, however, that it was from thermophilic ancestors that very high realized that these salt-loving invaded diverse low-temperature abundance “bacteria” are actually members environments. -

Archaea and the Origin of Eukaryotes
REVIEWS Archaea and the origin of eukaryotes Laura Eme, Anja Spang, Jonathan Lombard, Courtney W. Stairs and Thijs J. G. Ettema Abstract | Woese and Fox’s 1977 paper on the discovery of the Archaea triggered a revolution in the field of evolutionary biology by showing that life was divided into not only prokaryotes and eukaryotes. Rather, they revealed that prokaryotes comprise two distinct types of organisms, the Bacteria and the Archaea. In subsequent years, molecular phylogenetic analyses indicated that eukaryotes and the Archaea represent sister groups in the tree of life. During the genomic era, it became evident that eukaryotic cells possess a mixture of archaeal and bacterial features in addition to eukaryotic-specific features. Although it has been generally accepted for some time that mitochondria descend from endosymbiotic alphaproteobacteria, the precise evolutionary relationship between eukaryotes and archaea has continued to be a subject of debate. In this Review, we outline a brief history of the changing shape of the tree of life and examine how the recent discovery of a myriad of diverse archaeal lineages has changed our understanding of the evolutionary relationships between the three domains of life and the origin of eukaryotes. Furthermore, we revisit central questions regarding the process of eukaryogenesis and discuss what can currently be inferred about the evolutionary transition from the first to the last eukaryotic common ancestor. Sister groups Two descendants that split The pioneering work by Carl Woese and colleagues In this Review, we discuss how culture- independent from the same node; the revealed that all cellular life could be divided into three genomics has transformed our understanding of descendants are each other’s major evolutionary lines (also called domains): the archaeal diversity and how this has influenced our closest relative. -

Exploring the Cell Cycle of Archaea
Digital Comprehensive Summaries of Uppsala Dissertations from the Faculty of Science and Technology 300 Exploring the Cell Cycle of Archaea MAGNUS LUNDGREN ACTA UNIVERSITATIS UPSALIENSIS ISSN 1651-6214 UPPSALA ISBN 978-91-554-6881-1 2007 urn:nbn:se:uu:diva-7848 ! " # ! $%%& '( ) * * * + , - . , #, $%%&, / / * 0 , 0 , '%%, & , , 123 4&!54 5))657!! 5 , 0 * * , 1 * . , * * , - * * , . 5 * , " * . * 8 * 5 , 2 . 0 . . , 2 * 0 9 . * 5 , 0 * . , . * * . . , 0 , /7 . . 0 , -. /7 * . * , : . . * , 0 * ; . * / , 0 . . , < ; . * , - . . . 8 * , 1 , ! " 0 / : # / #$ % $ & $ ' ( )* $ $ %+,-./ $ ! = # $%%& 122 7) 57$ 6 123 4&!54 5))657!! 5 ( ((( 5&!6! > (?? ,,? @ A ( ((( 5&!6!B List of papers This thesis is based on the following papers, which are referred to in the text by their roman numerals. I Robinson NP, Dionne I*, Lundgren M*, Marsh VL, Bernander R, Bell SD. Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus. Cell, -

Bacteria and Archaea
DOMAINS AND KINGDOMS – Chapter 1 and 27-31 Archaea, Bacteria, Eukarya – Protista, including Algae, and Fungi Carl Woese based on studies of r-RNA of smaller ribosomal subunit in various groups of living things suggested 3-Domain classification of living things. The arrangement of nucleotides is highly conserved because the mutation rate in r-RNA is very slow. # BACTERIA ARCHAEA EUKARYA 1 Prokaryotic cells Prokaryotic cells Eukaryotic cells 2 Circular chromosome present Present Absent. Linear DNA in chromosomes 3 Peptidoglycan in cell wall present absent absent 4 No Histones associate with DNA Histones + DNA Histones + DNA 5 RNA polymerases: 1 type several types several types 6 Introns (non-coding part of gene) rare Sometimes present present 7 Membrane bound organelles absent absent Present Table 27.2 BACTERIA: multiple kingdoms Cell Wall: contains Peptidoglycan. It can get stained with crystal violet-Iodine. If the bacteria retain the stain on washing-these are called Gram+. If the stain is washed, the bacteria are stained with Safranin. These are called Gram- bacteria and have a second membrane outside cell wall. Forms: 3 main forms exist. 1 Bacillus - rod shaped bacteria 2 Coccus - spherical bacteria 3 Spiral or Curved bacteria Cell Structure: Bacteria lack all membrane bound organelles including nucleus. There are no histones associated with DNA. Ribosomes are smaller than ribosomes of eukaryotes. Flagella lack 9+2 arrangement. Metabolism: Bacteria are both autotrophs and heterotrophs. Most require organic molecules for source of energy and carbon source (chemoheterotrophs). These are saprobes and release enzymes to absorb food from outside. Besides fungi these are the main decomposers. -

Bacteria, Fungi and Archaea Domains in Rhizospheric Soil and Their Effects in Enhancing Agricultural Productivity
International Journal of Environmental Research and Public Health Review Bacteria, Fungi and Archaea Domains in Rhizospheric Soil and Their Effects in Enhancing Agricultural Productivity Kehinde Abraham Odelade and Olubukola Oluranti Babalola * Food Security and Safety Niche Area, Faculty of Natural and Agricultural Science, North-West University, Private Bag X2046, Mmabatho 2735, South Africa * Correspondence: [email protected]; Tel.: +27-18-389-2568 Received: 5 September 2019; Accepted: 4 October 2019; Published: 12 October 2019 Abstract: The persistent and undiscriminating application of chemicals as means to improve crop growth, development and yields for several years has become problematic to agricultural sustainability because of the adverse effects these chemicals have on the produce, consumers and beneficial microbes in the ecosystem. Therefore, for agricultural productivity to be sustained there are needs for better and suitable preferences which would be friendly to the ecosystem. The use of microbial metabolites has become an attractive and more feasible preference because they are versatile, degradable and ecofriendly, unlike chemicals. In order to achieve this aim, it is then imperative to explore microbes that are very close to the root of a plant, especially where they are more concentrated and have efficient activities called the rhizosphere. Extensive varieties of bacteria, archaea, fungi and other microbes are found inhabiting the rhizosphere with various interactions with the plant host. Therefore, this review explores various beneficial microbes such as bacteria, fungi and archaea and their roles in the environment in terms of acquisition of nutrients for plants for the purposes of plant growth and health. It also discusses the effect of root exudate on the rhizosphere microbiome and compares the three domains at molecular levels. -

Viruses and Prokaryotes in the Deep-Sea
Viruses and prokaryotes in the deep-sea Author: Roberto DANOVARO Professor of Marine Biology Polytechnic University of Marche, Ancona, Italy In the last decades the development of new technological and methodological tools has allowed demonstrating that microbes dominates both in terms of abundance and biomass the world oceans. Microbes are of size ranging from 0.02 to a few micrometres and include a wide diversity of viruses, prokaryotes (Bacteria and Archaea), and eukaryotes (either phototrophic and heterotrophic). These tiny particles, either suspended in the water column or attached to the sediments play crucially important functions and control the global biogeochemical cycles. More recently it has been also demonstrated that viruses are by far more important than previously thought. In particular we now know that viruses are the most abundant biological entities of the biosphere, with current estimates of the global viral abundance in the order of 1030-1031. They outnumber prokaryotes by at least one order of magnitude. This huge numerical abundance suggests that viruses can account also for the vast majority of the genetic diversity of the Earth (figure 1). Being not able to self-replicate, viruses invade other organisms and use their cell’s machinery to propagate. They can infect all the known life forms of the oceans, from the largest mammals to the smallest marine microbes. Indeed, also tiny viruses inside larger viruses have been recently discovered. Figure 1. Some examples of Viruses and Prokaryotes (i.e., Bacteria and Archaea) -

Osmoadaptation Mechanisms of Cyanobacteria and Archaea from the Stromatolites of Hamelin Pool, Western Australia
Osmoadaptation mechanisms of cyanobacteria and archaea from the stromatolites of Hamelin Pool, Western Australia Falicia Qi Yun Goh B. Sc (Hons.) UNSW A Dissertation Submitted in Fulfilment of the Requirements for the degree of Doctor of Philosophy School of Biotechnology and Biomolecular Sciences The University of New South Wales Sydney, Australia 2007 Osmoadaptation mechanisms of cyanobacteria and archaea from the stromatolites of Hamelin Pool, Western Australia Falicia Qi Yun Goh B. Sc (Hons.) (UNSW) A Dissertation Submitted in Partial Fulfilment of the Requirements for the degree of Doctor of Philosophy School of Biotechnology and Biomolecular Sciences The University of New South Wales Sydney, Australia Supervisors Prof. Brett A. Neilan Dr. Brendan P. Burns School of Biotechnology and Biomolecular Sciences The University of New South Wales Sydney, Australia Certificate of originality Abstract The stromatolites of Shark Bay Western Australia, located in a hypersaline environment, is an ideal biological system for studying survival strategies of cyanobacteria and halophilic archaea to high salt and their metabolic cooperation with other bacteria. To-date, little is known of the mechanisms by which these stromatolite microorganisms adapt to hypersalinity. To understand the formation of these sedimentary structures, detailed analysis of the microbial communities and their physiology for adaptation in this environment are crucial. In this study, microbial communities were investigated using culturing and molecular methods. Phylogenetic analysis of the 16S rRNA gene was carried out to investigate the diversity of microorganisms present. Unique phylotypes from the bacteria, cyanobacteria and archaea clone libraries were identified. Representative cyanobacteria isolates and Halococcus hamelinensis, a halophilic archaea isolated from in this study, were the focus for identifying osmoadaptation mechanisms. -

Archaea in Coastal Marine Environments (Achabaterla/Phyoey/Batwe Nt/N a Eclogu) EDWARD F
Proc. Natl. Acad. Sci. USA Vol. 89, pp. 5685-5689, June 1992 Ecology Archaea in coastal marine environments (achabaterla/phyoey/batwe nt/n a eclogU) EDWARD F. DELONG* Biology Department, Woods Hole Oceanographic Institution, Woods Hole, MA 02543 Communicated by George N. Somero, March 17, 1992 (receivedfor review February 4, 1992) ABSTRACT Archaea (archaebacteria) are a phenotypi- macroaggregate samples, eubacterial- or archaeal-biased cally diverse group of microorganisms that share a common PCR primers were routinely used to exclude the amplification evolutionary history. There are four general phenotypic groups of eukaryotic ribosomal RNA-encoding DNA (rDNA). Sur- of archaea: the methanogens, the extreme halophiles, the prisingly, archaeal rDNA was detected in many samples. sulfate-reducing archaea, and the extreme thermophiles. In the This report describes the detection of two marine archaeal marine environment, archaeal habitats are generally limited to lineages and their preliminary phylogenetic and ecological shallow or deep-sea anaerobic sediments (free-living and en- characterization. dosymbiotic methanogens), hot springs or deep-sea hydrother- mal vents (methanogens, sulfate reducers, and extreme ther- mophiles), and highly saline land-locked seas (halophiles). This METHODS report provides evidence for the widespread occurrence of Bacterioplankton Collection. Coastal water samples were unusual archaea in oxygenated coasl surface waters of North collected and screened through a 10-L&m Nytex mesh prefil- America. Quantitative mates indicated that up to 2% ofthe ter. Bacterioplankton were concentrated from these 10-pm- total ribosomal RNA extracted fom coastal bacterioplankton filtered water samples by using a CH2PR filtration unit assemblages was archaeal. Archaeal small-subunit ribosomal (Amicon) fitted with a polysulfone hollow-fiber filter (30-kDa RNA-encoding DNAs (rDNAs) were coned from mixed bac- cutoff).