Archaea and the Origin of Eukaryotes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Anoxygenic Photosynthesis in Photolithotrophic Sulfur Bacteria and Their Role in Detoxication of Hydrogen Sulfide
antioxidants Review Anoxygenic Photosynthesis in Photolithotrophic Sulfur Bacteria and Their Role in Detoxication of Hydrogen Sulfide Ivan Kushkevych 1,* , Veronika Bosáková 1,2 , Monika Vítˇezová 1 and Simon K.-M. R. Rittmann 3,* 1 Department of Experimental Biology, Faculty of Science, Masaryk University, 62500 Brno, Czech Republic; [email protected] (V.B.); [email protected] (M.V.) 2 Department of Biology, Faculty of Medicine, Masaryk University, 62500 Brno, Czech Republic 3 Archaea Physiology & Biotechnology Group, Department of Functional and Evolutionary Ecology, Universität Wien, 1090 Vienna, Austria * Correspondence: [email protected] (I.K.); [email protected] (S.K.-M.R.R.); Tel.: +420-549-495-315 (I.K.); +431-427-776-513 (S.K.-M.R.R.) Abstract: Hydrogen sulfide is a toxic compound that can affect various groups of water microorgan- isms. Photolithotrophic sulfur bacteria including Chromatiaceae and Chlorobiaceae are able to convert inorganic substrate (hydrogen sulfide and carbon dioxide) into organic matter deriving energy from photosynthesis. This process takes place in the absence of molecular oxygen and is referred to as anoxygenic photosynthesis, in which exogenous electron donors are needed. These donors may be reduced sulfur compounds such as hydrogen sulfide. This paper deals with the description of this metabolic process, representatives of the above-mentioned families, and discusses the possibility using anoxygenic phototrophic microorganisms for the detoxification of toxic hydrogen sulfide. Moreover, their general characteristics, morphology, metabolism, and taxonomy are described as Citation: Kushkevych, I.; Bosáková, well as the conditions for isolation and cultivation of these microorganisms will be presented. V.; Vítˇezová,M.; Rittmann, S.K.-M.R. -

Limits of Life on Earth Some Archaea and Bacteria
Limits of life on Earth Thermophiles Temperatures up to ~55C are common, but T > 55C is Some archaea and bacteria (extremophiles) can live in associated usually with geothermal features (hot springs, environments that we would consider inhospitable to volcanic activity etc) life (heat, cold, acidity, high pressure etc) Thermophiles are organisms that can successfully live Distinguish between growth and survival: many organisms can survive intervals of harsh conditions but could not at high temperatures live permanently in such conditions (e.g. seeds, spores) Best studied extremophiles: may be relevant to the Interest: origin of life. Very hot environments tolerable for life do not seem to exist elsewhere in the Solar System • analogs for extraterrestrial environments • `extreme’ conditions may have been more common on the early Earth - origin of life? • some unusual environments (e.g. subterranean) are very widespread Extraterrestrial Life: Spring 2008 Extraterrestrial Life: Spring 2008 Grand Prismatic Spring, Yellowstone National Park Hydrothermal vents: high pressure in the deep ocean allows liquid water Colors on the edge of the at T >> 100C spring are caused by different colonies of thermophilic Vents emit superheated water (300C or cyanobacteria and algae more) that is rich in minerals Hottest water is lifeless, but `cooler’ ~50 species of such thermophiles - mostly archae with some margins support array of thermophiles: cyanobacteria and anaerobic photosynthetic bacteria oxidize sulphur, manganese, grow on methane + carbon monoxide etc… Sulfolobus: optimum T ~ 80C, minimum 60C, maximum 90C, also prefer a moderately acidic pH. Live by oxidizing sulfur Known examples can grow (i.e. multiply) at temperatures which is abundant near hot springs. -

UV Laser-Induced, Time-Resolved Transcriptome Responses of Saccharomyces Cerevisiae
University of Tennessee, Knoxville TRACE: Tennessee Research and Creative Exchange Microbiology Publications and Other Works Microbiology 2019 UV Laser-Induced, Time-Resolved Transcriptome Responses of Saccharomyces cerevisiae Melinda Hauser University of Tennessee, Knoxville Paul E. Abraham Oak Ridge National Laboratory, Oak Ridge Lorenz Barcelona University of Tennessee, Knoxville Jeffrey M. Becker University of Tennessee, Knoxville Follow this and additional works at: https://trace.tennessee.edu/utk_micrpubs Recommended Citation Hauser, Melinda; Abraham, Paul E.; Barcelona, Lorenz; and Becker, Jeffrey M., "UV Laser-Induced, Time- Resolved Transcriptome Responses of Saccharomyces cerevisiae" (2019). Microbiology Publications and Other Works. https://trace.tennessee.edu/utk_micrpubs/107 This Article is brought to you for free and open access by the Microbiology at TRACE: Tennessee Research and Creative Exchange. It has been accepted for inclusion in Microbiology Publications and Other Works by an authorized administrator of TRACE: Tennessee Research and Creative Exchange. For more information, please contact [email protected]. INVESTIGATION UV Laser-Induced, Time-Resolved Transcriptome Responses of Saccharomyces cerevisiae Melinda Hauser,* Paul E. Abraham,† Lorenz Barcelona,*,1 and Jeffrey M. Becker*,2 *Department of Microbiology, University of Tennessee, Knoxville, TN 37996 and †Chemical Sciences Division, Oak Ridge National Laboratory, Oak Ridge, TN 37831 ORCID ID: 0000-0003-0467-5913 (J.M.B.) ABSTRACT We determined the effect on gene transcription of laser-mediated, long-wavelength KEYWORDS UV-irradiation of Saccharomyces cerevisiae by RNAseq analysis at times T15, T30, and T60 min after re- yeast covery in growth medium. Laser-irradiated cells were viable, and the transcriptional response was transient, gene expression with over 400 genes differentially expressed at T15 or T30, returning to basal level transcription by T60. -

The Genome of Nanoarchaeum Equitans: Insights Into Early Archaeal Evolution and Derived Parasitism
The genome of Nanoarchaeum equitans: Insights into early archaeal evolution and derived parasitism Elizabeth Waters†‡, Michael J. Hohn§, Ivan Ahel¶, David E. Graham††, Mark D. Adams‡‡, Mary Barnstead‡‡, Karen Y. Beeson‡‡, Lisa Bibbs†, Randall Bolanos‡‡, Martin Keller†, Keith Kretz†, Xiaoying Lin‡‡, Eric Mathur†, Jingwei Ni‡‡, Mircea Podar†, Toby Richardson†, Granger G. Sutton‡‡, Melvin Simon†, Dieter So¨ ll¶§§¶¶, Karl O. Stetter†§¶¶, Jay M. Short†, and Michiel Noordewier†¶¶ †Diversa Corporation, 4955 Directors Place, San Diego, CA 92121; ‡Department of Biology, San Diego State University, 5500 Campanile Drive, San Diego, CA 92182; §Lehrstuhl fu¨r Mikrobiologie und Archaeenzentrum, Universita¨t Regensburg, Universita¨tsstrasse 31, D-93053 Regensburg, Germany; ‡‡Celera Genomics Rockville, 45 West Gude Drive, Rockville, MD 20850; Departments of ¶Molecular Biophysics and Biochemistry and §§Chemistry, Yale University, New Haven, CT 06520-8114; and ʈDepartment of Biochemistry, Virginia Polytechnic Institute and State University, Blacksburg, VA 24061 Communicated by Carl R. Woese, University of Illinois at Urbana–Champaign, Urbana, IL, August 21, 2003 (received for review July 22, 2003) The hyperthermophile Nanoarchaeum equitans is an obligate sym- (6–8). Genomic DNA was either digested with restriction en- biont growing in coculture with the crenarchaeon Ignicoccus. zymes or sheared to provide clonable fragments. Two plasmid Ribosomal protein and rRNA-based phylogenies place its branching libraries were made by subcloning randomly sheared fragments point early in the archaeal lineage, representing the new archaeal of this DNA into a high-copy number vector (Ϸ2.8 kbp library) kingdom Nanoarchaeota. The N. equitans genome (490,885 base or low-copy number vector (Ϸ6.3 kbp library). DNA sequence pairs) encodes the machinery for information processing and was obtained from both ends of plasmid inserts to create repair, but lacks genes for lipid, cofactor, amino acid, or nucleotide ‘‘mate-pairs,’’ pairs of reads from single clones that should be biosyntheses. -

Two Domains of Life, Not Three?
Asgard Archaea Two domains of life, not three? A picture of visual interpretation of the Wagner’s opera Das Rheingold, a part of Richard Wagner’s Der Ring des Nibelungen Loki's Castle is a field of five active hydrothermal vents in the mid-Atlantic Ocean, located at 73 degrees north on the Mid-Atlantic Ridge between Greenland and Norway at a depth of 2,352 metres The vents were discovered in 2008 by a multinational scientific expedition of the university of Bergen, and are the most northerly black smokers to date. The five active chimneys of Loki's Castle are venting water as hot as 300 °C and sit on a vast mound of sulfide minerals. The vent field was given the name Loki's Castle as its shape reminded its discoverers of a fantasy castle. The reference is to the ancient Norse god of trickery, Loki. The top three feet (1 m) of a vent chimney almost 40 feet (12 m) tall at Loki's Castle in mid-July 2008. Visible at left is the arm of a remotely operated vehicle, reaching in to take fluid samples. Loki’s Castle Wents Loki's Castle - a field of five active hydrothermal vents in the mid-Atlantic Ocean - at 73 degrees north on the Mid-Atlantic Ridge - at a depth of 2,352 meters In Norse mythology, Loki is a cunning trickster who has the ability to change his shape and sex. Loki is represented as the companion of the great gods Odin and Thor. Preliminary observations have shown the warm area around the Loki's Castle vents to be alive with diverse and apparently unique microorganisms, unlike vent communities observed elsewhere. -

S41564-017-0083-5.Pdf
CORRECTION https://doi.org/10.1038/s41564-017-0083-5 Author Correction: Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life Donovan H. Parks, Christian Rinke, Maria Chuvochina, Pierre-Alain Chaumeil, Ben J. Woodcroft, Paul N. Evans, Philip Hugenholtz* and Gene W. Tyson* Correction to: Nature Microbiology https://doi.org/10.1038/s41564-017-0012-7 (2017); published online 11 September 2017. In the original version of this Article, the authors stated that the archaeal phylum Parvarchaeota was previously represented by only two single-cell genomes (ARMAN-4_'5-way FS' and ARMAN-5_'5-way FS'). However, these are in fact unpublished, low-quality metage- nome-assembled genomes (MAGs) obtained from Richmond Mine, California. In addition, the authors overlooked two higher-quality published Parvarchaeota MAGs from the same habitat, ARMAN-4 (ADCE00000000) and ARMAN-5 (ADHF00000000) (B. J. Baker et al., Proc. Natl Acad. Sci. USA 107, 8806–8811; 2010). The ARMAN-4 and ARMAN-5 MAGs are estimated to be 68.0% and 76.7% complete with 3.3% and 5.6% contamination, respectively, based on the archaeal-specific marker sets of CheckM. The 11 Parvarchaeota genomes identified in our study were obtained from different Richmond Mine metagenomes, but are highly similar to the ARMAN-4 (ANI of ~99.7%) and ARMAN-5 (ANI of ~99.6%) MAGs. The highest-quality uncultivated bacteria and archaea (UBA) MAGs with similarity to ARMAN-4 and ARMAN-5 are 82.5% and 83.3% complete with 0.9% and 1.9% contamination, respectively. The Parvarchaeota rep- resents only 0.23% of the archaeal genome tree and addition of the ARMAN-4 and ARMAN-5 MAGs do not change the conclusions of this Article, but do impact the phylogenetic gain for this phylum. -

University of Oklahoma Graduate College Development of High
UNIVERSITY OF OKLAHOMA GRADUATE COLLEGE DEVELOPMENT OF HIGH-THROUGHPUT EXPERIMENTAL AND COMPUTATIONAL TECHNOLOGIES FOR ANALYZING MICROBIAL FUNCTIONS AND INTERACTIONS IN ENVIRONMENTAL METAGENOMES A DISSERTATION SUBMITTED TO THE GRADUATE FACULTY in partial fulfillment of the requirements for the Degree of DOCTOR OF PHILOSOPHY By ZHOU SHI Norman, Oklahoma 2017 DEVELOPMENT OF HIGH-THROUGHPUT EXPERIMENTAL AND COMPUTATIONAL TECHNOLOGIES FOR ANALYZING MICROBIAL FUNCTIONS AND INTERACTIONS IN ENVIRONMENTAL METAGENOMES A DISSERTATION APPROVED FOR THE DEPARTMENT OF MICROBIOLOGY AND PLANT BIOLOGY BY ______________________________ Dr. Lee Krumholz, Chair ______________________________ Dr. Meijun Zhu ______________________________ Dr. Sridhar Radhakrishnan ______________________________ Dr. Yiqi Luo ______________________________ Dr. Jizhong Zhou © Copyright by ZHOU SHI 2017 All Rights Reserved. Dedication I dedicate this dissertation to my wife, Mengting Yuan, whom I met and married during the time period of this work and whose love, faith and support is the greatest gift that I can ever imagine in my life; to my son, Andrew Shi, who is always curious, energetic and wonderful, and is my lasting source of enlightenment and true happiness; to my parents, Yiping Shi and Aiping Zeng, whose selfless love and care transcended the distance and time, and carried me whenever I need; to my aunt, Cindy Shi, whose unwavering support and encouragement instilled in me the spirit of striving for excellence; to my friend, Su Xu, who boarded on the same plane with me to the U.S. seven years ago and then has witnessed all parts of this work as my best friend, and Dian Zou, who passed away in an accident during the time period of this work and is a friend I will forever miss. -
Lokiarchaeota: Biologists Discover 'Missing Link' Microorganism
Home About Us News Archive Copyright Privacy Policy Contact Us Newsletter RSS HOME ASTRONOMY SPACE EXPLORATION ARCHAEOLOGY PALEONTOLOGY BIOLOGY PHYSICS Lokiarchaeota: Biologists Discover ‘Missing Link’ Microorganism May 7, 2015 by Sci-News.com « PREVIOUS Published in A team of biologists, co-led by Dr Lionel Guy and Dr Thijs J. G. Ettema from Biology Uppsala University in Sweden, has discovered a new group of Tagged as microorganisms that represents an intermediate form in-between the Archaea simple cells of bacteria and the complex cell types of eukaryotes. Bacteria Eukaryote Lokiarchaeota Follow Like 16k Share Tweet 12 Like 58 41 You Might Like Bottlenose Dolphins Form Highly Complex Networks of Friends Rorqual Whales This false-color image shows a cell of thermophilic methanogenic archaea. Image credit: University of Have Unique California Museum of Paleontology. Stretchy Nerves In 1977, biochemist Dr Carl Woese and his colleagues at the University of Illinois described an entirely new group of organisms, the Archaea (originally found in extreme environments, such as hydrothermal vents and terrestrial hot springs). The scientists were studying relationships among the prokaryotes using DNA Extinction of sequences, and found that Archaea have distinct molecular characteristics World’s Largest separating them from bacteria as well as from eukaryotes. They proposed that Herbivores May Lead to Empty life can be divided into three domains: Eukaryota, Eubacteria, and Landscapes, Say Archaebacteria. Researchers Despite that archaeal cells were simple and small like bacteria, scientists found that Archaea were more closely related to organisms with complex cell types, a group collectively known as ‘eukaryotes.’ This observation has puzzled Sichuan Bush biologists for years. -

Genomic Analysis of Family UBA6911 (Group 18 Acidobacteria)
bioRxiv preprint doi: https://doi.org/10.1101/2021.04.09.439258; this version posted April 10, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 2 Genomic analysis of family UBA6911 (Group 18 3 Acidobacteria) expands the metabolic capacities of the 4 phylum and highlights adaptations to terrestrial habitats. 5 6 Archana Yadav1, Jenna C. Borrelli1, Mostafa S. Elshahed1, and Noha H. Youssef1* 7 8 1Department of Microbiology and Molecular Genetics, Oklahoma State University, Stillwater, 9 OK 10 *Correspondence: Noha H. Youssef: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/2021.04.09.439258; this version posted April 10, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 11 Abstract 12 Approaches for recovering and analyzing genomes belonging to novel, hitherto unexplored 13 bacterial lineages have provided invaluable insights into the metabolic capabilities and 14 ecological roles of yet-uncultured taxa. The phylum Acidobacteria is one of the most prevalent 15 and ecologically successful lineages on earth yet, currently, multiple lineages within this phylum 16 remain unexplored. Here, we utilize genomes recovered from Zodletone spring, an anaerobic 17 sulfide and sulfur-rich spring in southwestern Oklahoma, as well as from multiple disparate soil 18 and non-soil habitats, to examine the metabolic capabilities and ecological role of members of 19 the family UBA6911 (group18) Acidobacteria. -

Coupled Reductive and Oxidative Sulfur Cycling in the Phototrophic Plate of a Meromictic Lake T
Geobiology (2014), 12, 451–468 DOI: 10.1111/gbi.12092 Coupled reductive and oxidative sulfur cycling in the phototrophic plate of a meromictic lake T. L. HAMILTON,1 R. J. BOVEE,2 V. THIEL,3 S. R. SATTIN,2 W. MOHR,2 I. SCHAPERDOTH,1 K. VOGL,3 W. P. GILHOOLY III,4 T. W. LYONS,5 L. P. TOMSHO,3 S. C. SCHUSTER,3,6 J. OVERMANN,7 D. A. BRYANT,3,6,8 A. PEARSON2 AND J. L. MACALADY1 1Department of Geosciences, Penn State Astrobiology Research Center (PSARC), The Pennsylvania State University, University Park, PA, USA 2Department of Earth and Planetary Sciences, Harvard University, Cambridge, MA, USA 3Department of Biochemistry and Molecular Biology, The Pennsylvania State University, University Park, PA, USA 4Department of Earth Sciences, Indiana University-Purdue University Indianapolis, Indianapolis, IN, USA 5Department of Earth Sciences, University of California, Riverside, CA, USA 6Singapore Center for Environmental Life Sciences Engineering, Nanyang Technological University, Nanyang, Singapore 7Leibniz-Institut DSMZ-Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany 8Department of Chemistry and Biochemistry, Montana State University, Bozeman, MT, USA ABSTRACT Mahoney Lake represents an extreme meromictic model system and is a valuable site for examining the organisms and processes that sustain photic zone euxinia (PZE). A single population of purple sulfur bacte- ria (PSB) living in a dense phototrophic plate in the chemocline is responsible for most of the primary pro- duction in Mahoney Lake. Here, we present metagenomic data from this phototrophic plate – including the genome of the major PSB, as obtained from both a highly enriched culture and from the metagenomic data – as well as evidence for multiple other taxa that contribute to the oxidative sulfur cycle and to sulfate reduction. -

Yu-Chen Ling and John W. Moreau
Microbial Distribution and Activity in a Coastal Acid Sulfate Soil System Introduction: Bioremediation in Yu-Chen Ling and John W. Moreau coastal acid sulfate soil systems Method A Coastal acid sulfate soil (CASS) systems were School of Earth Sciences, University of Melbourne, Melbourne, VIC 3010, Australia formed when people drained the coastal area Microbial distribution controlled by environmental parameters Microbial activity showed two patterns exposing the soil to the air. Drainage makes iron Microbial structures can be grouped into three zones based on the highest similarity between samples (Fig. 4). Abundant populations, such as Deltaproteobacteria, kept constant activity across tidal cycling, whereas rare sulfides oxidize and release acidity to the These three zones were consistent with their geological background (Fig. 5). Zone 1: Organic horizon, had the populations changed activity response to environmental variations. Activity = cDNA/DNA environment, low pH pore water further dissolved lowest pH value. Zone 2: surface tidal zone, was influenced the most by tidal activity. Zone 3: Sulfuric zone, Abundant populations: the heavy metals. The acidity and toxic metals then Method A Deltaproteobacteria Deltaproteobacteria this area got neutralized the most. contaminate coastal and nearby ecosystems and Method B 1.5 cause environmental problems, such as fish kills, 1.5 decreased rice yields, release of greenhouse gases, Chloroflexi and construction damage. In Australia, there is Gammaproteobacteria Gammaproteobacteria about a $10 billion “legacy” from acid sulfate soils, Chloroflexi even though Australia is only occupied by around 1.0 1.0 Cyanobacteria,@ Acidobacteria Acidobacteria Alphaproteobacteria 18% of the global acid sulfate soils. Chloroplast Zetaproteobacteria Rare populations: Alphaproteobacteria Method A log(RNA(%)+1) Zetaproteobacteria log(RNA(%)+1) Method C Method B 0.5 0.5 Cyanobacteria,@ Bacteroidetes Chloroplast Firmicutes Firmicutes Bacteroidetes Planctomycetes Planctomycetes Ac8nobacteria Fig. -
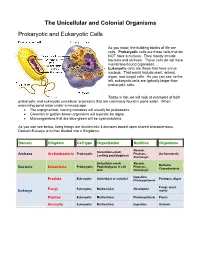
The Unicellular and Colonial Organisms Prokaryotic And
The Unicellular and Colonial Organisms Prokaryotic and Eukaryotic Cells As you know, the building blocks of life are cells. Prokaryotic cells are those cells that do NOT have a nucleus. They mostly include bacteria and archaea. These cells do not have membrane-bound organelles. Eukaryotic cells are those that have a true nucleus. That would include plant, animal, algae, and fungal cells. As you can see, to the left, eukaryotic cells are typically larger than prokaryotic cells. Today in lab, we will look at examples of both prokaryotic and eukaryotic unicellular organisms that are commonly found in pond water. When examining pond water under a microscope… The unpigmented, moving microbes will usually be protozoans. Greenish or golden-brown organisms will typically be algae. Microorganisms that are blue-green will be cyanobacteria. As you can see below, living things are divided into 3 domains based upon shared characteristics. Domain Eukarya is further divided into 4 Kingdoms. Domain Kingdom Cell type Organization Nutrition Organisms Absorb, Unicellular-small; Prokaryotic Photsyn., Archaeacteria Archaea Archaebacteria Lacking peptidoglycan Chemosyn. Unicellular-small; Absorb, Bacteria, Prokaryotic Peptidoglycan in cell Photsyn., Bacteria Eubacteria Cyanobacteria wall Chemosyn. Ingestion, Eukaryotic Unicellular or colonial Protozoa, Algae Protista Photosynthesis Fungi, yeast, Fungi Eukaryotic Multicellular Absorption Eukarya molds Plantae Eukaryotic Multicellular Photosynthesis Plants Animalia Eukaryotic Multicellular Ingestion Animals Prokaryotic Organisms – the archaea, non-photosynthetic bacteria, and cyanobacteria Archaea - Microorganisms that resemble bacteria, but are different from them in certain aspects. Archaea cell walls do not include the macromolecule peptidoglycan, which is always found in the cell walls of bacteria. Archaea usually live in extreme, often very hot or salty environments, such as hot mineral springs or deep-sea hydrothermal vents.