Exploring the Cell Cycle of Archaea
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Anoxygenic Photosynthesis in Photolithotrophic Sulfur Bacteria and Their Role in Detoxication of Hydrogen Sulfide
antioxidants Review Anoxygenic Photosynthesis in Photolithotrophic Sulfur Bacteria and Their Role in Detoxication of Hydrogen Sulfide Ivan Kushkevych 1,* , Veronika Bosáková 1,2 , Monika Vítˇezová 1 and Simon K.-M. R. Rittmann 3,* 1 Department of Experimental Biology, Faculty of Science, Masaryk University, 62500 Brno, Czech Republic; [email protected] (V.B.); [email protected] (M.V.) 2 Department of Biology, Faculty of Medicine, Masaryk University, 62500 Brno, Czech Republic 3 Archaea Physiology & Biotechnology Group, Department of Functional and Evolutionary Ecology, Universität Wien, 1090 Vienna, Austria * Correspondence: [email protected] (I.K.); [email protected] (S.K.-M.R.R.); Tel.: +420-549-495-315 (I.K.); +431-427-776-513 (S.K.-M.R.R.) Abstract: Hydrogen sulfide is a toxic compound that can affect various groups of water microorgan- isms. Photolithotrophic sulfur bacteria including Chromatiaceae and Chlorobiaceae are able to convert inorganic substrate (hydrogen sulfide and carbon dioxide) into organic matter deriving energy from photosynthesis. This process takes place in the absence of molecular oxygen and is referred to as anoxygenic photosynthesis, in which exogenous electron donors are needed. These donors may be reduced sulfur compounds such as hydrogen sulfide. This paper deals with the description of this metabolic process, representatives of the above-mentioned families, and discusses the possibility using anoxygenic phototrophic microorganisms for the detoxification of toxic hydrogen sulfide. Moreover, their general characteristics, morphology, metabolism, and taxonomy are described as Citation: Kushkevych, I.; Bosáková, well as the conditions for isolation and cultivation of these microorganisms will be presented. V.; Vítˇezová,M.; Rittmann, S.K.-M.R. -

Limits of Life on Earth Some Archaea and Bacteria
Limits of life on Earth Thermophiles Temperatures up to ~55C are common, but T > 55C is Some archaea and bacteria (extremophiles) can live in associated usually with geothermal features (hot springs, environments that we would consider inhospitable to volcanic activity etc) life (heat, cold, acidity, high pressure etc) Thermophiles are organisms that can successfully live Distinguish between growth and survival: many organisms can survive intervals of harsh conditions but could not at high temperatures live permanently in such conditions (e.g. seeds, spores) Best studied extremophiles: may be relevant to the Interest: origin of life. Very hot environments tolerable for life do not seem to exist elsewhere in the Solar System • analogs for extraterrestrial environments • `extreme’ conditions may have been more common on the early Earth - origin of life? • some unusual environments (e.g. subterranean) are very widespread Extraterrestrial Life: Spring 2008 Extraterrestrial Life: Spring 2008 Grand Prismatic Spring, Yellowstone National Park Hydrothermal vents: high pressure in the deep ocean allows liquid water Colors on the edge of the at T >> 100C spring are caused by different colonies of thermophilic Vents emit superheated water (300C or cyanobacteria and algae more) that is rich in minerals Hottest water is lifeless, but `cooler’ ~50 species of such thermophiles - mostly archae with some margins support array of thermophiles: cyanobacteria and anaerobic photosynthetic bacteria oxidize sulphur, manganese, grow on methane + carbon monoxide etc… Sulfolobus: optimum T ~ 80C, minimum 60C, maximum 90C, also prefer a moderately acidic pH. Live by oxidizing sulfur Known examples can grow (i.e. multiply) at temperatures which is abundant near hot springs. -

Extreme Organisms on Earth Show Us Just How Weird Life Elsewhere Could Be. by Chris Impey Astrobiology
Astrobiology Extreme organisms on Earth show us just how weird life elsewhere could be. by Chris Impey How life could thrive on hostile worlds Humans have left their mark all over Earth. We’re proud of our role as nature’s generalists — perhaps not as swift as the gazelle or as strong as the gorilla, but still pretty good at most things. Alone among all species, technology has given us dominion over the planet. Humans are endlessly plucky and adaptable; it seems we can do anything. Strain 121 Yet in truth, we’re frail. From our safe living rooms, we may admire the people who conquer Everest or cross deserts. But without technology, we couldn’t live beyond Earth’s temperate zones. We cannot survive for long in temperatures below freezing or above 104° Fahrenheit (40° Celsius). We can stay underwater only as long as we can hold our breath. Without water to drink we’d die in 3 days. Microbes, on the other hand, are hardy. And within the microbial world lies a band of extremists, organisms that thrive in conditions that would cook, crush, smother, and dissolve most other forms of life. Collectively, they are known as extremophiles, which means, literally, “lovers of extremes.” Extremophiles are found at temperatures above the boiling point and below the freezing point of water, in high salinity, and in strongly acidic conditions. Some can live deep inside rock, and others can go into a freeze-dried “wait state” for tens of thousands of years. Some of these microbes harvest energy from meth- ane, sulfur, and even iron. -
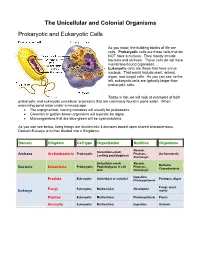
The Unicellular and Colonial Organisms Prokaryotic And
The Unicellular and Colonial Organisms Prokaryotic and Eukaryotic Cells As you know, the building blocks of life are cells. Prokaryotic cells are those cells that do NOT have a nucleus. They mostly include bacteria and archaea. These cells do not have membrane-bound organelles. Eukaryotic cells are those that have a true nucleus. That would include plant, animal, algae, and fungal cells. As you can see, to the left, eukaryotic cells are typically larger than prokaryotic cells. Today in lab, we will look at examples of both prokaryotic and eukaryotic unicellular organisms that are commonly found in pond water. When examining pond water under a microscope… The unpigmented, moving microbes will usually be protozoans. Greenish or golden-brown organisms will typically be algae. Microorganisms that are blue-green will be cyanobacteria. As you can see below, living things are divided into 3 domains based upon shared characteristics. Domain Eukarya is further divided into 4 Kingdoms. Domain Kingdom Cell type Organization Nutrition Organisms Absorb, Unicellular-small; Prokaryotic Photsyn., Archaeacteria Archaea Archaebacteria Lacking peptidoglycan Chemosyn. Unicellular-small; Absorb, Bacteria, Prokaryotic Peptidoglycan in cell Photsyn., Bacteria Eubacteria Cyanobacteria wall Chemosyn. Ingestion, Eukaryotic Unicellular or colonial Protozoa, Algae Protista Photosynthesis Fungi, yeast, Fungi Eukaryotic Multicellular Absorption Eukarya molds Plantae Eukaryotic Multicellular Photosynthesis Plants Animalia Eukaryotic Multicellular Ingestion Animals Prokaryotic Organisms – the archaea, non-photosynthetic bacteria, and cyanobacteria Archaea - Microorganisms that resemble bacteria, but are different from them in certain aspects. Archaea cell walls do not include the macromolecule peptidoglycan, which is always found in the cell walls of bacteria. Archaea usually live in extreme, often very hot or salty environments, such as hot mineral springs or deep-sea hydrothermal vents. -

Archaeal Distribution and Abundance in Water Masses of the Arctic Ocean, Pacific Sector
Vol. 69: 101–112, 2013 AQUATIC MICROBIAL ECOLOGY Published online April 30 doi: 10.3354/ame01624 Aquat Microb Ecol FREEREE ACCESSCCESS Archaeal distribution and abundance in water masses of the Arctic Ocean, Pacific sector Chie Amano-Sato1, Shohei Akiyama1, Masao Uchida2, Koji Shimada3, Motoo Utsumi1,* 1University of Tsukuba, Tennodai, Tsukuba, Ibaraki 305-8572, Japan 2National Institute for Environmental Studies, Onogawa, Tsukuba, Ibaraki 305-8506, Japan 3Tokyo University of Marine Science and Technology, Konan, Minato-ku, Tokyo 108-8477, Japan ABSTRACT: Marine planktonic Archaea have been recently recognized as an ecologically impor- tant component of marine prokaryotic biomass in the world’s oceans. Their abundance and meta- bolism are closely connected with marine geochemical cycling. We evaluated the distribution of planktonic Archaea in the Pacific sector of the Arctic Ocean using fluorescence in situ hybridiza- tion (FISH) with catalyzed reporter deposition (CARD-FISH) and performed statistical analyses using data for archaeal abundance and geochemical variables. The relative abundance of Thaum - archaeota generally increased with depth, and euryarchaeal abundance was the lowest of all planktonic prokaryotes. Multiple regression analysis showed that the thaumarchaeal relative abundance was negatively correlated with ammonium and dissolved oxygen concentrations and chlorophyll fluorescence. Canonical correspondence analysis showed that archaeal distributions differed with oceanographic water masses; in particular, Thaumarchaeota were abundant from the halocline layer to deep water, where salinity was higher and most nutrients were depleted. However, at several stations on the East Siberian Sea side of the study area and along the North- wind Ridge, Thaumarchaeota and Bacteria were proportionally very abundant at the bottom in association with higher nutrient conditions. -

Oceans of Archaea Abundant Oceanic Crenarchaeota Appear to Derive from Thermophilic Ancestors That Invaded Low-Temperature Marine Environments
Oceans of Archaea Abundant oceanic Crenarchaeota appear to derive from thermophilic ancestors that invaded low-temperature marine environments Edward F. DeLong arth’s microbiota is remarkably per- karyotes), Archaea, and Bacteria. Although al- vasive, thriving at extremely high ternative taxonomic schemes have been recently temperature, low and high pH, high proposed, whole-genome and other analyses E salinity, and low water availability. tend to support Woese’s three-domain concept. One lineage of microbial life in par- Well-known and cultivated archaea generally ticular, the Archaea, is especially adept at ex- fall into several major phenotypic groupings: ploiting environmental extremes. Despite their these include extreme halophiles, methanogens, success in these challenging habitats, the Ar- and extreme thermophiles and thermoacido- chaea may now also be viewed as a philes. Early on, extremely halo- cosmopolitan lot. These microbes philic archaea (haloarchaea) were exist in a wide variety of terres- first noticed as bright-red colonies trial, freshwater, and marine habi- Archaea exist in growing on salted fish or hides. tats, sometimes in very high abun- a wide variety For many years, halophilic isolates dance. The oceanic Marine Group of terrestrial, from salterns, salt deposits, and I Crenarchaeota, for example, ri- freshwater, and landlocked seas provided excellent val total bacterial biomass in wa- marine habitats, model systems for studying adap- ters below 100 m. These wide- tations to high salinity. It was only spread Archaea appear to derive sometimes in much later, however, that it was from thermophilic ancestors that very high realized that these salt-loving invaded diverse low-temperature abundance “bacteria” are actually members environments. -

Roles of Methylation and Sequestration in the Mechanisms of DNA Replication in Some Members of the Enterobacteriaceae Family
Chapter 12 Roles of Methylation and Sequestration in the Mechanisms of DNA Replication in some Members of the Enterobacteriaceae Family Amine Aloui, Alya El May, Saloua Kouass Sahbani and Ahmed Landoulsi Additional information is available at the end of the chapter http://dx.doi.org/10.5772/51724 1. Introduction When growing cells divide, they need to copy their genetic material and distribute it to en‐ sure that each daughter cell receives one copy. This is a challenging task especially when the enormous length of the DNA compared to the cell size is considered. During DNA replica‐ tion, organization of the chromosomes is even more demanding, since replication forks con‐ tinuously produce new DNA. This DNA contains all the information required to build the cells and tissues of a prokaryotic or an eukaryotic organism. The exact replication of this in‐ formation in any species assures its genetic continuity from generation to generation and is critical to the normal development of an individual. The information stored in DNA is ar‐ ranged in hereditary units known as genes that control the identifiable traits of an organism. Discovery of the structure of DNA and subsequent elucidation of how DNA directs synthe‐ sis of RNA, which then directs assembly of proteins -the so-called central dogma - were monumental achievements that marked the early days of molecular biology. However, the simplified representation of the central dogma as DNA → RNA → protein does not reflect the role of proteins in the synthesis of nucleic acids. Moreover, proteins are largely responsi‐ ble for regulating DNA replication and gene expression, the entire process whereby the in‐ formation encoded in DNA is decoded into the proteins that characterize various cell types. -

Archaea and the Origin of Eukaryotes
REVIEWS Archaea and the origin of eukaryotes Laura Eme, Anja Spang, Jonathan Lombard, Courtney W. Stairs and Thijs J. G. Ettema Abstract | Woese and Fox’s 1977 paper on the discovery of the Archaea triggered a revolution in the field of evolutionary biology by showing that life was divided into not only prokaryotes and eukaryotes. Rather, they revealed that prokaryotes comprise two distinct types of organisms, the Bacteria and the Archaea. In subsequent years, molecular phylogenetic analyses indicated that eukaryotes and the Archaea represent sister groups in the tree of life. During the genomic era, it became evident that eukaryotic cells possess a mixture of archaeal and bacterial features in addition to eukaryotic-specific features. Although it has been generally accepted for some time that mitochondria descend from endosymbiotic alphaproteobacteria, the precise evolutionary relationship between eukaryotes and archaea has continued to be a subject of debate. In this Review, we outline a brief history of the changing shape of the tree of life and examine how the recent discovery of a myriad of diverse archaeal lineages has changed our understanding of the evolutionary relationships between the three domains of life and the origin of eukaryotes. Furthermore, we revisit central questions regarding the process of eukaryogenesis and discuss what can currently be inferred about the evolutionary transition from the first to the last eukaryotic common ancestor. Sister groups Two descendants that split The pioneering work by Carl Woese and colleagues In this Review, we discuss how culture- independent from the same node; the revealed that all cellular life could be divided into three genomics has transformed our understanding of descendants are each other’s major evolutionary lines (also called domains): the archaeal diversity and how this has influenced our closest relative. -

The Thermal Limits to Life on Earth
International Journal of Astrobiology 13 (2): 141–154 (2014) doi:10.1017/S1473550413000438 © Cambridge University Press 2014. The online version of this article is published within an Open Access environment subject to the conditions of the Creative Commons Attribution licence http://creativecommons.org/licenses/by/3.0/. The thermal limits to life on Earth Andrew Clarke1,2 1British Antarctic Survey, Cambridge, UK 2School of Environmental Sciences, University of East Anglia, Norwich, UK e-mail: [email protected] Abstract: Living organisms on Earth are characterized by three necessary features: a set of internal instructions encoded in DNA (software), a suite of proteins and associated macromolecules providing a boundary and internal structure (hardware), and a flux of energy. In addition, they replicate themselves through reproduction, a process that renders evolutionary change inevitable in a resource-limited world. Temperature has a profound effect on all of these features, and yet life is sufficiently adaptable to be found almost everywhere water is liquid. The thermal limits to survival are well documented for many types of organisms, but the thermal limits to completion of the life cycle are much more difficult to establish, especially for organisms that inhabit thermally variable environments. Current data suggest that the thermal limits to completion of the life cycle differ between the three major domains of life, bacteria, archaea and eukaryotes. At the very highest temperatures only archaea are found with the current high-temperature limit for growth being 122 °C. Bacteria can grow up to 100 °C, but no eukaryote appears to be able to complete its life cycle above *60 °C and most not above 40 °C. -

Negative Control of Replication Initiation by a Novel Chromosomal Locus Exhibiting Exceptional Affinity for Escherichia Coli Dnaa Protein
Downloaded from genesdev.cshlp.org on October 2, 2021 - Published by Cold Spring Harbor Laboratory Press Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein Risa Kitagawa,1,3 Toru Ozaki,1 Shigeki Moriya,2 and Tohru Ogawa1,4 1Division of Biological Science, Graduate School of Science, Nagoya University Chikusa-ku, Nagoya 464-8602, Japan; 2Department of Cell Biology, Graduate School of Biological Sciences, Nara Institute of Science and Technology, Takayama-cho, Ikoma 630-0101, Japan Replication of the Escherichia coli chromosome is initiated at a unique site, oriC. Concurrent initiation occurs at all oriC sites present in a cell once, and only once, per cell cycle. A mechanism to ensure cyclic initiation events was found operating through the chromosomal site, datA, a 1-kb segment located at 94.7 min on the genetic map that titrates exceptionally large amounts of the bacterial initiator protein, DnaA. A strain lacking datA grew normally but exhibited an asynchronous initiation phenotype as a result of extra initiation events. This mutant phenotype was suppressed by DnaA-titrating plasmids. Furthermore, mutations in a 9-bp DnaA-binding sequence (the DnaA box) in datA were enough to induce the mutant phenotype. Thus, datA is a novel chromosomal element that appears to adjust a balance between free and bound DnaA for a single initiation event at a fixed time in the bacterial cell cycle. Titration of DnaA to newly duplicated datA during oriC sequestration, which is mediated by hemimethylated GATC sequences in oriC and the SeqA protein, would contribute to prevention of reinitiations when oriC is desequestered. -

Microbial Extremophiles in Aspect of Limits of Life. Elena V. ~Ikuta
Source of Acquisition NASA Marshall Space Flight Centel Microbial extremophiles in aspect of limits of life. Elena V. ~ikuta',Richard B. ~oover*,and Jane an^.^ IT LF " '-~ationalSpace Sciences and Technology CenterINASA, VP-62, 320 Sparkman Dr., Astrobiology Laboratory, Huntsville, AL 35805, USA. 3- Noblis, 3 150 Fairview Park Drive South, Falls Church, VA 22042, USA. Introduction. During Earth's evolution accompanied by geophysical and climatic changes a number of ecosystems have been formed. These ecosystems differ by the broad variety of physicochemical and biological factors composing our environment. Traditionally, pH and salinity are considered as geochemical extremes, as opposed to the temperature, pressure and radiation that are referred to as physical extremes (Van den Burg, 2003). Life inhabits all possible places on Earth interacting with the environment and within itself (cross species relations). In nature it is very rare when an ecotope is inhabited by a single species. As a rule, most ecosystems contain the functionally related and evolutionarily adjusted communities (consortia and populations). In contrast to the multicellular structure of eukaryotes (tissues, organs, systems of organs, whole organism), the highest organized form of prokaryotic life in nature is the benthic colonization in biofilms and microbial mats. In these complex structures all microbial cells of different species are distributed in space and time according to their functions and to physicochemical gradients that allow more effective system support, self- protection, and energy distribution. In vitro, of course, the most primitive organized structure for bacterial and archaeal cultures is the colony, the size, shape, color, consistency, and other characteristics of which could carry varies specifics on species or subspecies levels. -

Post-Genomic Characterization of Metabolic Pathways in Sulfolobus Solfataricus
Post-Genomic Characterization of Metabolic Pathways in Sulfolobus solfataricus Jasper Walther Thesis committee Thesis supervisors Prof. dr. J. van der Oost Personal chair at the laboratory of Microbiology Wageningen University Prof. dr. W. M. de Vos Professor of Microbiology Wageningen University Other members Prof. dr. W.J.H. van Berkel Wageningen University Prof. dr. V.A.F. Martins dos Santos Wageningen University Dr. T.J.G. Ettema Uppsala University, Sweden Dr. S.V. Albers Max Planck Institute for Terrestrial Microbiology, Marburg, Germany This research was conducted under the auspices of the Graduate School VLAG Post-Genomic Characterization of Metabolic Pathways in Sulfolobus solfataricus Jasper Walther Thesis Submitted in fulfilment of the requirements for the degree of doctor at Wageningen University by the authority of the Rector Magnificus Prof. dr. M.J. Kropff, in the presence of the Thesis Committee appointed by the Academic Board to be defended in public on Monday 23 January 2012 at 11 a.m. in the Aula. Jasper Walther Post-Genomic Characterization of Metabolic Pathways in Sulfolobus solfataricus, 164 pages. Thesis, Wageningen University, Wageningen, NL (2012) With references, with summaries in Dutch and English ISBN 978-94-6173-203-3 Table of contents Chapter 1 Introduction 1 Chapter 2 Hot Transcriptomics 17 Chapter 3 Reconstruction of central carbon metabolism in Sulfolobus solfataricus using a two-dimensional gel electrophoresis map, stable isotope labelling and DNA microarray analysis 45 Chapter 4 Identification of the Missing