The Origin of Eukaryotes and Their Relationship with the Archaea: Are
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Anoxygenic Photosynthesis in Photolithotrophic Sulfur Bacteria and Their Role in Detoxication of Hydrogen Sulfide
antioxidants Review Anoxygenic Photosynthesis in Photolithotrophic Sulfur Bacteria and Their Role in Detoxication of Hydrogen Sulfide Ivan Kushkevych 1,* , Veronika Bosáková 1,2 , Monika Vítˇezová 1 and Simon K.-M. R. Rittmann 3,* 1 Department of Experimental Biology, Faculty of Science, Masaryk University, 62500 Brno, Czech Republic; [email protected] (V.B.); [email protected] (M.V.) 2 Department of Biology, Faculty of Medicine, Masaryk University, 62500 Brno, Czech Republic 3 Archaea Physiology & Biotechnology Group, Department of Functional and Evolutionary Ecology, Universität Wien, 1090 Vienna, Austria * Correspondence: [email protected] (I.K.); [email protected] (S.K.-M.R.R.); Tel.: +420-549-495-315 (I.K.); +431-427-776-513 (S.K.-M.R.R.) Abstract: Hydrogen sulfide is a toxic compound that can affect various groups of water microorgan- isms. Photolithotrophic sulfur bacteria including Chromatiaceae and Chlorobiaceae are able to convert inorganic substrate (hydrogen sulfide and carbon dioxide) into organic matter deriving energy from photosynthesis. This process takes place in the absence of molecular oxygen and is referred to as anoxygenic photosynthesis, in which exogenous electron donors are needed. These donors may be reduced sulfur compounds such as hydrogen sulfide. This paper deals with the description of this metabolic process, representatives of the above-mentioned families, and discusses the possibility using anoxygenic phototrophic microorganisms for the detoxification of toxic hydrogen sulfide. Moreover, their general characteristics, morphology, metabolism, and taxonomy are described as Citation: Kushkevych, I.; Bosáková, well as the conditions for isolation and cultivation of these microorganisms will be presented. V.; Vítˇezová,M.; Rittmann, S.K.-M.R. -

Limits of Life on Earth Some Archaea and Bacteria
Limits of life on Earth Thermophiles Temperatures up to ~55C are common, but T > 55C is Some archaea and bacteria (extremophiles) can live in associated usually with geothermal features (hot springs, environments that we would consider inhospitable to volcanic activity etc) life (heat, cold, acidity, high pressure etc) Thermophiles are organisms that can successfully live Distinguish between growth and survival: many organisms can survive intervals of harsh conditions but could not at high temperatures live permanently in such conditions (e.g. seeds, spores) Best studied extremophiles: may be relevant to the Interest: origin of life. Very hot environments tolerable for life do not seem to exist elsewhere in the Solar System • analogs for extraterrestrial environments • `extreme’ conditions may have been more common on the early Earth - origin of life? • some unusual environments (e.g. subterranean) are very widespread Extraterrestrial Life: Spring 2008 Extraterrestrial Life: Spring 2008 Grand Prismatic Spring, Yellowstone National Park Hydrothermal vents: high pressure in the deep ocean allows liquid water Colors on the edge of the at T >> 100C spring are caused by different colonies of thermophilic Vents emit superheated water (300C or cyanobacteria and algae more) that is rich in minerals Hottest water is lifeless, but `cooler’ ~50 species of such thermophiles - mostly archae with some margins support array of thermophiles: cyanobacteria and anaerobic photosynthetic bacteria oxidize sulphur, manganese, grow on methane + carbon monoxide etc… Sulfolobus: optimum T ~ 80C, minimum 60C, maximum 90C, also prefer a moderately acidic pH. Live by oxidizing sulfur Known examples can grow (i.e. multiply) at temperatures which is abundant near hot springs. -

Genomic Analysis of Family UBA6911 (Group 18 Acidobacteria)
bioRxiv preprint doi: https://doi.org/10.1101/2021.04.09.439258; this version posted April 10, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 2 Genomic analysis of family UBA6911 (Group 18 3 Acidobacteria) expands the metabolic capacities of the 4 phylum and highlights adaptations to terrestrial habitats. 5 6 Archana Yadav1, Jenna C. Borrelli1, Mostafa S. Elshahed1, and Noha H. Youssef1* 7 8 1Department of Microbiology and Molecular Genetics, Oklahoma State University, Stillwater, 9 OK 10 *Correspondence: Noha H. Youssef: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/2021.04.09.439258; this version posted April 10, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 11 Abstract 12 Approaches for recovering and analyzing genomes belonging to novel, hitherto unexplored 13 bacterial lineages have provided invaluable insights into the metabolic capabilities and 14 ecological roles of yet-uncultured taxa. The phylum Acidobacteria is one of the most prevalent 15 and ecologically successful lineages on earth yet, currently, multiple lineages within this phylum 16 remain unexplored. Here, we utilize genomes recovered from Zodletone spring, an anaerobic 17 sulfide and sulfur-rich spring in southwestern Oklahoma, as well as from multiple disparate soil 18 and non-soil habitats, to examine the metabolic capabilities and ecological role of members of 19 the family UBA6911 (group18) Acidobacteria. -

Coupled Reductive and Oxidative Sulfur Cycling in the Phototrophic Plate of a Meromictic Lake T
Geobiology (2014), 12, 451–468 DOI: 10.1111/gbi.12092 Coupled reductive and oxidative sulfur cycling in the phototrophic plate of a meromictic lake T. L. HAMILTON,1 R. J. BOVEE,2 V. THIEL,3 S. R. SATTIN,2 W. MOHR,2 I. SCHAPERDOTH,1 K. VOGL,3 W. P. GILHOOLY III,4 T. W. LYONS,5 L. P. TOMSHO,3 S. C. SCHUSTER,3,6 J. OVERMANN,7 D. A. BRYANT,3,6,8 A. PEARSON2 AND J. L. MACALADY1 1Department of Geosciences, Penn State Astrobiology Research Center (PSARC), The Pennsylvania State University, University Park, PA, USA 2Department of Earth and Planetary Sciences, Harvard University, Cambridge, MA, USA 3Department of Biochemistry and Molecular Biology, The Pennsylvania State University, University Park, PA, USA 4Department of Earth Sciences, Indiana University-Purdue University Indianapolis, Indianapolis, IN, USA 5Department of Earth Sciences, University of California, Riverside, CA, USA 6Singapore Center for Environmental Life Sciences Engineering, Nanyang Technological University, Nanyang, Singapore 7Leibniz-Institut DSMZ-Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany 8Department of Chemistry and Biochemistry, Montana State University, Bozeman, MT, USA ABSTRACT Mahoney Lake represents an extreme meromictic model system and is a valuable site for examining the organisms and processes that sustain photic zone euxinia (PZE). A single population of purple sulfur bacte- ria (PSB) living in a dense phototrophic plate in the chemocline is responsible for most of the primary pro- duction in Mahoney Lake. Here, we present metagenomic data from this phototrophic plate – including the genome of the major PSB, as obtained from both a highly enriched culture and from the metagenomic data – as well as evidence for multiple other taxa that contribute to the oxidative sulfur cycle and to sulfate reduction. -

Yu-Chen Ling and John W. Moreau
Microbial Distribution and Activity in a Coastal Acid Sulfate Soil System Introduction: Bioremediation in Yu-Chen Ling and John W. Moreau coastal acid sulfate soil systems Method A Coastal acid sulfate soil (CASS) systems were School of Earth Sciences, University of Melbourne, Melbourne, VIC 3010, Australia formed when people drained the coastal area Microbial distribution controlled by environmental parameters Microbial activity showed two patterns exposing the soil to the air. Drainage makes iron Microbial structures can be grouped into three zones based on the highest similarity between samples (Fig. 4). Abundant populations, such as Deltaproteobacteria, kept constant activity across tidal cycling, whereas rare sulfides oxidize and release acidity to the These three zones were consistent with their geological background (Fig. 5). Zone 1: Organic horizon, had the populations changed activity response to environmental variations. Activity = cDNA/DNA environment, low pH pore water further dissolved lowest pH value. Zone 2: surface tidal zone, was influenced the most by tidal activity. Zone 3: Sulfuric zone, Abundant populations: the heavy metals. The acidity and toxic metals then Method A Deltaproteobacteria Deltaproteobacteria this area got neutralized the most. contaminate coastal and nearby ecosystems and Method B 1.5 cause environmental problems, such as fish kills, 1.5 decreased rice yields, release of greenhouse gases, Chloroflexi and construction damage. In Australia, there is Gammaproteobacteria Gammaproteobacteria about a $10 billion “legacy” from acid sulfate soils, Chloroflexi even though Australia is only occupied by around 1.0 1.0 Cyanobacteria,@ Acidobacteria Acidobacteria Alphaproteobacteria 18% of the global acid sulfate soils. Chloroplast Zetaproteobacteria Rare populations: Alphaproteobacteria Method A log(RNA(%)+1) Zetaproteobacteria log(RNA(%)+1) Method C Method B 0.5 0.5 Cyanobacteria,@ Bacteroidetes Chloroplast Firmicutes Firmicutes Bacteroidetes Planctomycetes Planctomycetes Ac8nobacteria Fig. -
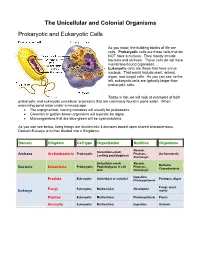
The Unicellular and Colonial Organisms Prokaryotic And
The Unicellular and Colonial Organisms Prokaryotic and Eukaryotic Cells As you know, the building blocks of life are cells. Prokaryotic cells are those cells that do NOT have a nucleus. They mostly include bacteria and archaea. These cells do not have membrane-bound organelles. Eukaryotic cells are those that have a true nucleus. That would include plant, animal, algae, and fungal cells. As you can see, to the left, eukaryotic cells are typically larger than prokaryotic cells. Today in lab, we will look at examples of both prokaryotic and eukaryotic unicellular organisms that are commonly found in pond water. When examining pond water under a microscope… The unpigmented, moving microbes will usually be protozoans. Greenish or golden-brown organisms will typically be algae. Microorganisms that are blue-green will be cyanobacteria. As you can see below, living things are divided into 3 domains based upon shared characteristics. Domain Eukarya is further divided into 4 Kingdoms. Domain Kingdom Cell type Organization Nutrition Organisms Absorb, Unicellular-small; Prokaryotic Photsyn., Archaeacteria Archaea Archaebacteria Lacking peptidoglycan Chemosyn. Unicellular-small; Absorb, Bacteria, Prokaryotic Peptidoglycan in cell Photsyn., Bacteria Eubacteria Cyanobacteria wall Chemosyn. Ingestion, Eukaryotic Unicellular or colonial Protozoa, Algae Protista Photosynthesis Fungi, yeast, Fungi Eukaryotic Multicellular Absorption Eukarya molds Plantae Eukaryotic Multicellular Photosynthesis Plants Animalia Eukaryotic Multicellular Ingestion Animals Prokaryotic Organisms – the archaea, non-photosynthetic bacteria, and cyanobacteria Archaea - Microorganisms that resemble bacteria, but are different from them in certain aspects. Archaea cell walls do not include the macromolecule peptidoglycan, which is always found in the cell walls of bacteria. Archaea usually live in extreme, often very hot or salty environments, such as hot mineral springs or deep-sea hydrothermal vents. -

Archaeal Distribution and Abundance in Water Masses of the Arctic Ocean, Pacific Sector
Vol. 69: 101–112, 2013 AQUATIC MICROBIAL ECOLOGY Published online April 30 doi: 10.3354/ame01624 Aquat Microb Ecol FREEREE ACCESSCCESS Archaeal distribution and abundance in water masses of the Arctic Ocean, Pacific sector Chie Amano-Sato1, Shohei Akiyama1, Masao Uchida2, Koji Shimada3, Motoo Utsumi1,* 1University of Tsukuba, Tennodai, Tsukuba, Ibaraki 305-8572, Japan 2National Institute for Environmental Studies, Onogawa, Tsukuba, Ibaraki 305-8506, Japan 3Tokyo University of Marine Science and Technology, Konan, Minato-ku, Tokyo 108-8477, Japan ABSTRACT: Marine planktonic Archaea have been recently recognized as an ecologically impor- tant component of marine prokaryotic biomass in the world’s oceans. Their abundance and meta- bolism are closely connected with marine geochemical cycling. We evaluated the distribution of planktonic Archaea in the Pacific sector of the Arctic Ocean using fluorescence in situ hybridiza- tion (FISH) with catalyzed reporter deposition (CARD-FISH) and performed statistical analyses using data for archaeal abundance and geochemical variables. The relative abundance of Thaum - archaeota generally increased with depth, and euryarchaeal abundance was the lowest of all planktonic prokaryotes. Multiple regression analysis showed that the thaumarchaeal relative abundance was negatively correlated with ammonium and dissolved oxygen concentrations and chlorophyll fluorescence. Canonical correspondence analysis showed that archaeal distributions differed with oceanographic water masses; in particular, Thaumarchaeota were abundant from the halocline layer to deep water, where salinity was higher and most nutrients were depleted. However, at several stations on the East Siberian Sea side of the study area and along the North- wind Ridge, Thaumarchaeota and Bacteria were proportionally very abundant at the bottom in association with higher nutrient conditions. -

Insights Into Archaeal Evolution and Symbiosis from the Genomes of a Nanoarchaeon and Its Inferred Crenarchaeal Host from Obsidian Pool, Yellowstone National Park
University of Tennessee, Knoxville TRACE: Tennessee Research and Creative Exchange Microbiology Publications and Other Works Microbiology 4-22-2013 Insights into archaeal evolution and symbiosis from the genomes of a nanoarchaeon and its inferred crenarchaeal host from Obsidian Pool, Yellowstone National Park Mircea Podar University of Tennessee - Knoxville, [email protected] Kira S. Makarova National Institutes of Health David E. Graham University of Tennessee - Knoxville, [email protected] Yuri I. Wolf National Institutes of Health Eugene V. Koonin National Institutes of Health See next page for additional authors Follow this and additional works at: https://trace.tennessee.edu/utk_micrpubs Part of the Microbiology Commons Recommended Citation Biology Direct 2013, 8:9 doi:10.1186/1745-6150-8-9 This Article is brought to you for free and open access by the Microbiology at TRACE: Tennessee Research and Creative Exchange. It has been accepted for inclusion in Microbiology Publications and Other Works by an authorized administrator of TRACE: Tennessee Research and Creative Exchange. For more information, please contact [email protected]. Authors Mircea Podar, Kira S. Makarova, David E. Graham, Yuri I. Wolf, Eugene V. Koonin, and Anna-Louise Reysenbach This article is available at TRACE: Tennessee Research and Creative Exchange: https://trace.tennessee.edu/ utk_micrpubs/44 Podar et al. Biology Direct 2013, 8:9 http://www.biology-direct.com/content/8/1/9 RESEARCH Open Access Insights into archaeal evolution and symbiosis from the genomes of a nanoarchaeon and its inferred crenarchaeal host from Obsidian Pool, Yellowstone National Park Mircea Podar1,2*, Kira S Makarova3, David E Graham1,2, Yuri I Wolf3, Eugene V Koonin3 and Anna-Louise Reysenbach4 Abstract Background: A single cultured marine organism, Nanoarchaeum equitans, represents the Nanoarchaeota branch of symbiotic Archaea, with a highly reduced genome and unusual features such as multiple split genes. -

Oceans of Archaea Abundant Oceanic Crenarchaeota Appear to Derive from Thermophilic Ancestors That Invaded Low-Temperature Marine Environments
Oceans of Archaea Abundant oceanic Crenarchaeota appear to derive from thermophilic ancestors that invaded low-temperature marine environments Edward F. DeLong arth’s microbiota is remarkably per- karyotes), Archaea, and Bacteria. Although al- vasive, thriving at extremely high ternative taxonomic schemes have been recently temperature, low and high pH, high proposed, whole-genome and other analyses E salinity, and low water availability. tend to support Woese’s three-domain concept. One lineage of microbial life in par- Well-known and cultivated archaea generally ticular, the Archaea, is especially adept at ex- fall into several major phenotypic groupings: ploiting environmental extremes. Despite their these include extreme halophiles, methanogens, success in these challenging habitats, the Ar- and extreme thermophiles and thermoacido- chaea may now also be viewed as a philes. Early on, extremely halo- cosmopolitan lot. These microbes philic archaea (haloarchaea) were exist in a wide variety of terres- first noticed as bright-red colonies trial, freshwater, and marine habi- Archaea exist in growing on salted fish or hides. tats, sometimes in very high abun- a wide variety For many years, halophilic isolates dance. The oceanic Marine Group of terrestrial, from salterns, salt deposits, and I Crenarchaeota, for example, ri- freshwater, and landlocked seas provided excellent val total bacterial biomass in wa- marine habitats, model systems for studying adap- ters below 100 m. These wide- tations to high salinity. It was only spread Archaea appear to derive sometimes in much later, however, that it was from thermophilic ancestors that very high realized that these salt-loving invaded diverse low-temperature abundance “bacteria” are actually members environments. -

Origin and Evolution of Eukaryotic Compartmentalization
TESI DOCTORAL UPF 2016 Origin and evolution of eukaryotic compartmentalization Alexandros Pittis Director Dr. Toni Gabaldon´ Comparative Genomics Group Bioinformatics and Genomics Department Centre for Genomic Regulation (CRG) To my father Stavros for the PhD he never did Acknowledgments Looking back at the years of my PhD in the Comparative Genomics group at CRG, I feel it was equally a process of scientific, as well as personal development. All this time I was very privileged to be surrounded by people that offered me their generous support in both fronts. And they were available in the very moments that I was fighting more myself than the unsolvable (anyway) comparative genomics puzzles. I hope that in the future I will have many times the chance to express them my appreciation, way beyond these few words. First, to Toni, my supervisor, for all his support, and patience, and confidence, and respect, especially at the moments that things did not seem that promising. He offered me freedom, to try, to think, to fail, to learn, to achieve that few enjoy during their PhD, and I am very thankful to him. Then, to my good friends and colleagues, those that I found in the group already, and others that joined after me. A very special thanks to Marinita and Jaime, for all their valuable time, and guidance and paradigm, which nevertheless I never managed to follow. They have both marked my phylogenomics path so far and I cannot escape. To Les and Salvi, my fellow students and pals at the time for all that we shared; to Gab and Fran and Dam, for -

Novel Abundant Oceanic Viruses of Uncultured Marine Group II Euryarchaeota Identified by Genome-Centric Metagenomics
bioRxiv preprint doi: https://doi.org/10.1101/101196; this version posted January 18, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Novel Abundant Oceanic Viruses of Uncultured Marine Group II Euryarchaeota Identified by Genome-Centric Metagenomics Alon Philosof1*, Natalya Yutin2, José Flores-Uribe1, Itai Sharon3, Eugene V. Koonin2, 5 and Oded Béjà1* 1Faculty of Biology, Technion - Israel Institute of Technology, Haifa, Israel. 2National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD, 20894, USA. 10 3Migal Galilee Research Institute, Kiryat Shmona, 11016, Israel. Tel Hai College, Upper Galilee 12210, Israel. *To whom correspondence should be addressed. E-mail: [email protected] and E-mail: [email protected] 15 bioRxiv preprint doi: https://doi.org/10.1101/101196; this version posted January 18, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Marine Group II Euryarchaeota (MGII) are among the most abundant microbes in the oceanic surface waters. So far, however, representatives of MGII have not been 20 cultivated, and no viruses infecting these organisms have been described. Here we present complete genomes for 3 distinct groups of viruses assembled from metagenomic sequence datasets highly enriched for MGII. These novel viruses, which we denote Magroviruses, possess double-stranded DNA genomes of 65 to 100 kilobase in size that encode a structural module characteristic of head-tailed 25 viruses and, unusually for archaeal and bacterial viruses, a nearly complete replication apparatus of apparent archaeal origin. -

Radical Views of the Tree of Life
Environmental Microbiology (2009) doi:10.1111/j.1462-2920.2009.01895.x Genomics update Radical views of the Tree of Life Howard Ochman* been posited over the years (Embley and Martin, 2006; Department of Biochemistry and Molecular Biophysics, Poole and Penny, 2007). University of Arizona, Tucson, Arizona 85718, USA. To examine the origin of the eukaryotes, Pisani et al. (2007) applied a supertree approach to each of three The tripartite division of cellular organisms into three large overlapping data sets containing either 0, 8 or 21 domains – the Bacteria, the Archaea and the Eukarya – eukaryotic genomes, in order to monitor the effects of has become so ingrained that most of us have all but taxon sampling on the resulting branching orders. Super- forgotten (or did not even know) that the ancestry and trees are phylogenies that are assembled by merging sets relationships among these ancient lineages are far from of more limited trees, each based on a gene that is not settled. The most widely accepted view, which emerged necessarily present in all taxa. Their results resolved bac- primarily from analyses of anciently duplicated genes teria and archaea as separate groups, but favour a chi- (Gogarten et al., 1989; Iwabe et al., 1989), places Bacte- meric origin of eukaryotes, as the majority of eukaryotic ria as the basal lineage and distinct from the common genes with prokaryotic homologues derive from multiple ancestor of archaea and eukaryotes. Accordingly, these symbioses. These findings help explain how different studies frame the Archaea, despite their appellation, as genes originated and why particular genes show distinct archaic but not primordial, and the three domains are phylogenetic histories, but they do not ascertain whether each viewed as separate lineages, with archaea and the cellular lineage leading to eukaryotes – i.e.