Interactions Between Anaerobic Fungi and Methanogens in the Rumen and Their Biotechnological Potential in Biogas Production from Lignocellulosic Materials
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Anoxygenic Photosynthesis in Photolithotrophic Sulfur Bacteria and Their Role in Detoxication of Hydrogen Sulfide
antioxidants Review Anoxygenic Photosynthesis in Photolithotrophic Sulfur Bacteria and Their Role in Detoxication of Hydrogen Sulfide Ivan Kushkevych 1,* , Veronika Bosáková 1,2 , Monika Vítˇezová 1 and Simon K.-M. R. Rittmann 3,* 1 Department of Experimental Biology, Faculty of Science, Masaryk University, 62500 Brno, Czech Republic; [email protected] (V.B.); [email protected] (M.V.) 2 Department of Biology, Faculty of Medicine, Masaryk University, 62500 Brno, Czech Republic 3 Archaea Physiology & Biotechnology Group, Department of Functional and Evolutionary Ecology, Universität Wien, 1090 Vienna, Austria * Correspondence: [email protected] (I.K.); [email protected] (S.K.-M.R.R.); Tel.: +420-549-495-315 (I.K.); +431-427-776-513 (S.K.-M.R.R.) Abstract: Hydrogen sulfide is a toxic compound that can affect various groups of water microorgan- isms. Photolithotrophic sulfur bacteria including Chromatiaceae and Chlorobiaceae are able to convert inorganic substrate (hydrogen sulfide and carbon dioxide) into organic matter deriving energy from photosynthesis. This process takes place in the absence of molecular oxygen and is referred to as anoxygenic photosynthesis, in which exogenous electron donors are needed. These donors may be reduced sulfur compounds such as hydrogen sulfide. This paper deals with the description of this metabolic process, representatives of the above-mentioned families, and discusses the possibility using anoxygenic phototrophic microorganisms for the detoxification of toxic hydrogen sulfide. Moreover, their general characteristics, morphology, metabolism, and taxonomy are described as Citation: Kushkevych, I.; Bosáková, well as the conditions for isolation and cultivation of these microorganisms will be presented. V.; Vítˇezová,M.; Rittmann, S.K.-M.R. -

Complete Genome Sequence of Methanocorpusculum Labreanum Type Strain Z
Standards in Genomic Sciences (2009) 1: 197-203 DOI:10.4056.sigs.35575 Complete genome sequence of Methanocorpusculum labreanum type strain Z Iain J. Anderson1*, Magdalena Sieprawska-Lupa2, Eugene Goltsman1, Alla Lapidus1, Alex Co- peland1, Tijana Glavina Del Rio1, Hope Tice1, Eileen Dalin1, Kerrie Barry1, Sam Pitluck1, Lo- ren Hauser1,3, Miriam Land1,3, Susan Lucas1, Paul Richardson1, William B. Whitman2, and Nikos C. Kyrpides1 1Joint Genome Institute, 2800 Mitchell Drive, Walnut Creek, California, USA 2Microbiology Department, University of Georgia, Athens, Georgia, USA 3Oak Ridge National Laboratory, Oak Ridge, Tennessee, USA *Corresponding author: Iain Anderson Keywords: archaea, methanogen, Methanomicrobiales Methanocorpusculum labreanum is a methanogen belonging to the order Methanomicro- biales within the archaeal phylum Euryarchaeota. The type strain Z was isolated from surface sediments of Tar Pit Lake in the La Brea Tar Pits in Los Angeles, California. M. labreanum is of phylogenetic interest because at the time the sequencing project began only one genome had previously been sequenced from the order Methanomicrobiales. We report here the complete genome sequence of M. labreanum type strain Z and its annotation. This is part of a 2006 Joint Genome Institute Community Sequencing Program project to sequence genomes of diverse Archaea. Introduction Methanocorpusculum labreanum is a methanogen the Methanosarcinales are capable of using various belonging to the order Methanomicrobiales within methyl compounds as substrates for methanoge- the archaeal phylum Euryarchaeota. Strain Z is the nesis including acetate, methylamines, and me- type strain of this species. It was isolated from thanol, but Methanomicrobiales are restricted to surface sediments of Tar Pit Lake at the La Brea the same substrates as the Class I methanogens Tar Pits in Los Angeles [1]. -

Genome-Resolved Meta-Analysis of the Microbiome in Oil Reservoirs Worldwide
microorganisms Article Genome-Resolved Meta-Analysis of the Microbiome in Oil Reservoirs Worldwide Kelly J. Hidalgo 1,2,* , Isabel N. Sierra-Garcia 3 , German Zafra 4 and Valéria M. de Oliveira 1 1 Microbial Resources Division, Research Center for Chemistry, Biology and Agriculture (CPQBA), University of Campinas–UNICAMP, Av. Alexandre Cazellato 999, 13148-218 Paulínia, Brazil; [email protected] 2 Graduate Program in Genetics and Molecular Biology, Institute of Biology, University of Campinas (UNICAMP), Rua Monteiro Lobato 255, Cidade Universitária, 13083-862 Campinas, Brazil 3 Biology Department & CESAM, University of Aveiro, Aveiro, Portugal, Campus de Santiago, Avenida João Jacinto de Magalhães, 3810-193 Aveiro, Portugal; [email protected] 4 Grupo de Investigación en Bioquímica y Microbiología (GIBIM), Escuela de Microbiología, Universidad Industrial de Santander, Cra 27 calle 9, 680002 Bucaramanga, Colombia; [email protected] * Correspondence: [email protected]; Tel.: +55-19981721510 Abstract: Microorganisms inhabiting subsurface petroleum reservoirs are key players in biochemical transformations. The interactions of microbial communities in these environments are highly complex and still poorly understood. This work aimed to assess publicly available metagenomes from oil reservoirs and implement a robust pipeline of genome-resolved metagenomics to decipher metabolic and taxonomic profiles of petroleum reservoirs worldwide. Analysis of 301.2 Gb of metagenomic information derived from heavily flooded petroleum reservoirs in China and Alaska to non-flooded petroleum reservoirs in Brazil enabled us to reconstruct 148 metagenome-assembled genomes (MAGs) of high and medium quality. At the phylum level, 74% of MAGs belonged to bacteria and 26% to archaea. The profiles of these MAGs were related to the physicochemical parameters and recovery management applied. -

Review Article: Inhibition of Methanogenic Archaea by Statins As a Targeted Management Strategy for Constipation and Related Disorders
Alimentary Pharmacology and Therapeutics Review article: inhibition of methanogenic archaea by statins as a targeted management strategy for constipation and related disorders K. Gottlieb*, V. Wacher*, J. Sliman* & M. Pimentel† *Synthetic Biologics, Inc., Rockville, SUMMARY MD, USA. † Gastroenterology, Cedars-Sinai Background Medical Center, Los Angeles, CA, USA. Observational studies show a strong association between delayed intestinal transit and the production of methane. Experimental data suggest a direct inhibitory activity of methane on the colonic and ileal smooth muscle and Correspondence to: a possible role for methane as a gasotransmitter. Archaea are the only con- Dr K. Gottlieb, Synthetic Biologics, fi Inc., 9605 Medical Center Drive, rmed biological sources of methane in nature and Methanobrevibacter Rockville, MD 20850, USA. smithii is the predominant methanogen in the human intestine. E-mail: [email protected] Aim To review the biosynthesis and composition of archaeal cell membranes, Publication data archaeal methanogenesis and the mechanism of action of statins in this context. Submitted 8 September 2015 First decision 29 September 2015 Methods Resubmitted 7 October 2015 Narrative review of the literature. Resubmitted 20 October 2015 Accepted 20 October 2015 Results EV Pub Online 11 November 2015 Statins can inhibit archaeal cell membrane biosynthesis without affecting This uncommissioned review article was bacterial numbers as demonstrated in livestock and humans. This opens subject to full peer-review. the possibility of a therapeutic intervention that targets a specific aetiologi- cal factor of constipation while protecting the intestinal microbiome. While it is generally believed that statins inhibit methane production via their effect on cell membrane biosynthesis, mediated by inhibition of the HMG- CoA reductase, there is accumulating evidence for an alternative or addi- tional mechanism of action where statins inhibit methanogenesis directly. -

Regeneration of Unconventional Natural Gas by Methanogens Co
www.nature.com/scientificreports OPEN Regeneration of unconventional natural gas by methanogens co‑existing with sulfate‑reducing prokaryotes in deep shale wells in China Yimeng Zhang1,2,3, Zhisheng Yu1*, Yiming Zhang4 & Hongxun Zhang1 Biogenic methane in shallow shale reservoirs has been proven to contribute to economic recovery of unconventional natural gas. However, whether the microbes inhabiting the deeper shale reservoirs at an average depth of 4.1 km and even co-occurring with sulfate-reducing prokaryote (SRP) have the potential to produce biomethane is still unclear. Stable isotopic technique with culture‑dependent and independent approaches were employed to investigate the microbial and functional diversity related to methanogenic pathways and explore the relationship between SRP and methanogens in the shales in the Sichuan Basin, China. Although stable isotopic ratios of the gas implied a thermogenic origin for methane, the decreased trend of stable carbon and hydrogen isotope value provided clues for increasing microbial activities along with sustained gas production in these wells. These deep shale-gas wells harbored high abundance of methanogens (17.2%) with ability of utilizing various substrates for methanogenesis, which co-existed with SRP (6.7%). All genes required for performing methylotrophic, hydrogenotrophic and acetoclastic methanogenesis were present. Methane production experiments of produced water, with and without additional available substrates for methanogens, further confrmed biomethane production via all three methanogenic pathways. Statistical analysis and incubation tests revealed the partnership between SRP and methanogens under in situ sulfate concentration (~ 9 mg/L). These results suggest that biomethane could be produced with more fexible stimulation strategies for unconventional natural gas recovery even at the higher depths and at the presence of SRP. -

Limits of Life on Earth Some Archaea and Bacteria
Limits of life on Earth Thermophiles Temperatures up to ~55C are common, but T > 55C is Some archaea and bacteria (extremophiles) can live in associated usually with geothermal features (hot springs, environments that we would consider inhospitable to volcanic activity etc) life (heat, cold, acidity, high pressure etc) Thermophiles are organisms that can successfully live Distinguish between growth and survival: many organisms can survive intervals of harsh conditions but could not at high temperatures live permanently in such conditions (e.g. seeds, spores) Best studied extremophiles: may be relevant to the Interest: origin of life. Very hot environments tolerable for life do not seem to exist elsewhere in the Solar System • analogs for extraterrestrial environments • `extreme’ conditions may have been more common on the early Earth - origin of life? • some unusual environments (e.g. subterranean) are very widespread Extraterrestrial Life: Spring 2008 Extraterrestrial Life: Spring 2008 Grand Prismatic Spring, Yellowstone National Park Hydrothermal vents: high pressure in the deep ocean allows liquid water Colors on the edge of the at T >> 100C spring are caused by different colonies of thermophilic Vents emit superheated water (300C or cyanobacteria and algae more) that is rich in minerals Hottest water is lifeless, but `cooler’ ~50 species of such thermophiles - mostly archae with some margins support array of thermophiles: cyanobacteria and anaerobic photosynthetic bacteria oxidize sulphur, manganese, grow on methane + carbon monoxide etc… Sulfolobus: optimum T ~ 80C, minimum 60C, maximum 90C, also prefer a moderately acidic pH. Live by oxidizing sulfur Known examples can grow (i.e. multiply) at temperatures which is abundant near hot springs. -

Core Sulphate-Reducing Microorganisms in Metal-Removing Semi-Passive Biochemical Reactors and the Co-Occurrence of Methanogens
microorganisms Article Core Sulphate-Reducing Microorganisms in Metal-Removing Semi-Passive Biochemical Reactors and the Co-Occurrence of Methanogens Maryam Rezadehbashi and Susan A. Baldwin * Chemical and Biological Engineering, University of British Columbia, 2360 East Mall, Vancouver, BC V6T 1Z3, Canada; [email protected] * Correspondence: [email protected]; Tel.: +1-604-822-1973 Received: 2 January 2018; Accepted: 17 February 2018; Published: 23 February 2018 Abstract: Biochemical reactors (BCRs) based on the stimulation of sulphate-reducing microorganisms (SRM) are emerging semi-passive remediation technologies for treatment of mine-influenced water. Their successful removal of metals and sulphate has been proven at the pilot-scale, but little is known about the types of SRM that grow in these systems and whether they are diverse or restricted to particular phylogenetic or taxonomic groups. A phylogenetic study of four established pilot-scale BCRs on three different mine sites compared the diversity of SRM growing in them. The mine sites were geographically distant from each other, nevertheless the BCRs selected for similar SRM types. Clostridia SRM related to Desulfosporosinus spp. known to be tolerant to high concentrations of copper were members of the core microbial community. Members of the SRM family Desulfobacteraceae were dominant, particularly those related to Desulfatirhabdium butyrativorans. Methanogens were dominant archaea and possibly were present at higher relative abundances than SRM in some BCRs. Both hydrogenotrophic and acetoclastic types were present. There were no strong negative or positive co-occurrence correlations of methanogen and SRM taxa. Knowing which SRM inhabit successfully operating BCRs allows practitioners to target these phylogenetic groups when selecting inoculum for future operations. -

Application to Develop Low Risk Gmos
NO3P Develop in containment a project of low risk genetically ER-AF-NO3P-3 modified organisms by rapid assessment 12/07 Application title: Identification and characterization of Potential Methane Mitigation Technologies Applicant organisation: AgResearch Ltd Considered by: IBSC ERMA Please clearly identify any confidential information and attach as a separate appendix. Please complete the following before submitting your application: All sections completed Yes Appendices enclosed Yes/NA Confidential information identified and enclosed separately Yes/NA Copies of references attached Yes/NA Application signed and dated Yes Electronic copy of application e-mailed to ERMA New Yes Zealand Signed: Date: 9 May 2011 20 Customhouse Quay Cnr Waring Taylor and Customhouse Quay PO Box 131, Wellington Phone: 04 916 2426 Fax: 04 914 0433 Email: [email protected] Website: www.ermanz.govt.nz Develop in containment a project of low risk genetically modified organisms by rapid assessment 1. An associated User Guide NO3P is available for this form and we strongly advise that you read this User Guide before filling out this application form. If you need guidance in completing this form please contact ERMA New Zealand or your IBSC. 2. This application form only covers the development of low-risk genetically modified organisms that meet Category A and/or B experiments as defined in the HSNO (Low-Risk Genetic Modification) Regulations 2003. 3. If you are making an application that includes not low-risk genetic modification experiments, as described in the HSNO (Low-Risk Genetic Modification) Regulations 2003, then you should complete form NO3O instead. 4. This form replaces all previous versions of Form NO3P. -

Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline
Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline • Phlogenetics of Archaea • Phlogenetics of archaeal lipids • Papers Phyla • Two? main phyla – Euryarchaeota • Methanogens • Extreme halophiles • Extreme thermophiles • Sulfate-reducing – Crenarchaeota • Extreme thermophiles – Korarchaeota? • Hyperthermophiles • indicated only by environmental DNA sequences – Nanoarchaeum? • N. equitans a fast evolving euryarchaeal lineage, not novel, early diverging archaeal phylum – Ancient archael group? • In deepest brances of Crenarchaea? Euryarchaea? Archaeal Lipids • Methanogens – Di- and tetra-ethers of glycerol and isoprenoid alcohols – Core mostly archaeol or caldarchaeol – Core sometimes sn-2- or Images removed due to sn-3-hydroxyarchaeol or copyright considerations. macrocyclic archaeol –PMI • Halophiles – Similar to methanogens – Exclusively synthesize bacterioruberin • Marine Crenarchaea Depositional Archaeal Lipids Biological Origin Environment Crocetane methanotrophs? methane seeps? methanogens, PMI (2,6,10,15,19-pentamethylicosane) methanotrophs hypersaline, anoxic Squalane hypersaline? C31-C40 head-to-head isoprenoids Smit & Mushegian • “Lost” enzymes of MVA pathway must exist – Phosphomevalonate kinase (PMK) – Diphosphomevalonate decarboxylase – Isopentenyl diphosphate isomerase (IPPI) Kaneda et al. 2001 Rohdich et al. 2001 Boucher et al. • Isoprenoid biosynthesis of archaea evolved through a combination of processes – Co-option of ancestral enzymes – Modification of enzymatic specificity – Orthologous and non-orthologous gene -

Anaerobic Digestion of the Microalga Spirulina at Extreme Alkaline Conditions: Biogas Production, Metagenome, and Metatranscriptome
ORIGINAL RESEARCH published: 22 June 2015 doi: 10.3389/fmicb.2015.00597 Anaerobic digestion of the microalga Spirulina at extreme alkaline conditions: biogas production, metagenome, and metatranscriptome Vímac Nolla-Ardèvol 1*, Marc Strous 1, 2, 3 and Halina E. Tegetmeyer 1, 3, 4 1 Institute for Genome Research and Systems Biology, Center for Biotechnology, University of Bielefeld, Bielefeld, Germany, 2 Department of Geoscience, University of Calgary, Calgary, AB, Canada, 3 Microbial Fitness Group, Max Planck Institute for Marine Microbiology, Bremen, Germany, 4 HGF-MPG Group for Deep Sea Ecology and Technology, Alfred Wegener Institute, Helmholtz Centre for Polar and Marine Research, Bremerhaven, Germany A haloalkaline anaerobic microbial community obtained from soda lake sediments was Edited by: Mark Alexander Lever, used to inoculate anaerobic reactors for the production of methane rich biogas. The ETH Zürich, Switzerland microalga Spirulina was successfully digested by the haloalkaline microbial consortium + Reviewed by: at alkaline conditions (pH 10, 2.0 M Na ). Continuous biogas production was observed Aharon Oren, and the obtained biogas was rich in methane, up to 96%. Alkaline medium acted The Hebrew University of Jerusalem, Israel as a CO2 scrubber which resulted in low amounts of CO2 and no traces of H2S Ronald Oremland, in the produced biogas. A hydraulic retention time (HRT) of 15 days and 0.25 g United States Geological Survey, USA Spirulina L−1 day−1 organic loading rate (OLR) were identified as the optimal operational *Correspondence: Vímac Nolla-Ardèvol, parameters. Metagenomic and metatranscriptomic analysis showed that the hydrolysis Institute for Genome Research and of the supplied substrate was mainly carried out by Bacteroidetes of the “ML635J-40 Systems Biology, Center for aquatic group” while the hydrogenotrophic pathway was the main producer of methane Biotechnology, University of Bielefeld, Office G2-152, Universitätstraße 27, in a methanogenic community dominated by Methanocalculus. -

Supporting Information
Supporting Information Lozupone et al. 10.1073/pnas.0807339105 SI Methods nococcus, and Eubacterium grouped with members of other Determining the Environmental Distribution of Sequenced Genomes. named genera with high bootstrap support (Fig. 1A). One To obtain information on the lifestyle of the isolate and its reported member of the Bacteroidetes (Bacteroides capillosus) source, we looked at descriptive information from NCBI grouped firmly within the Firmicutes. This taxonomic error was (www.ncbi.nlm.nih.gov/genomes/lproks.cgi) and other related not surprising because gut isolates have often been classified as publications. We also determined which 16S rRNA-based envi- Bacteroides based on an obligate anaerobe, Gram-negative, ronmental surveys of microbial assemblages deposited near- nonsporulating phenotype alone (6, 7). A more recent 16S identical sequences in GenBank. We first downloaded the gbenv rRNA-based analysis of the genus Clostridium defined phylo- files from the NCBI ftp site on December 31, 2007, and used genetically related clusters (4, 5), and these designations were them to create a BLAST database. These files contain GenBank supported in our phylogenetic analysis of the Clostridium species in the HGMI pipeline. We thus designated these Clostridium records for the ENV database, a component of the nonredun- species, along with the species from other named genera that dant nucleotide database (nt) where 16S rRNA environmental cluster with them in bootstrap supported nodes, as being within survey data are deposited. GenBank records for hits with Ͼ98% these clusters. sequence identity over 400 bp to the 16S rRNA sequence of each of the 67 genomes were parsed to get a list of study titles Annotation of GTs and GHs. -
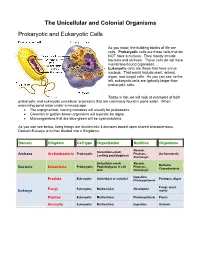
The Unicellular and Colonial Organisms Prokaryotic And
The Unicellular and Colonial Organisms Prokaryotic and Eukaryotic Cells As you know, the building blocks of life are cells. Prokaryotic cells are those cells that do NOT have a nucleus. They mostly include bacteria and archaea. These cells do not have membrane-bound organelles. Eukaryotic cells are those that have a true nucleus. That would include plant, animal, algae, and fungal cells. As you can see, to the left, eukaryotic cells are typically larger than prokaryotic cells. Today in lab, we will look at examples of both prokaryotic and eukaryotic unicellular organisms that are commonly found in pond water. When examining pond water under a microscope… The unpigmented, moving microbes will usually be protozoans. Greenish or golden-brown organisms will typically be algae. Microorganisms that are blue-green will be cyanobacteria. As you can see below, living things are divided into 3 domains based upon shared characteristics. Domain Eukarya is further divided into 4 Kingdoms. Domain Kingdom Cell type Organization Nutrition Organisms Absorb, Unicellular-small; Prokaryotic Photsyn., Archaeacteria Archaea Archaebacteria Lacking peptidoglycan Chemosyn. Unicellular-small; Absorb, Bacteria, Prokaryotic Peptidoglycan in cell Photsyn., Bacteria Eubacteria Cyanobacteria wall Chemosyn. Ingestion, Eukaryotic Unicellular or colonial Protozoa, Algae Protista Photosynthesis Fungi, yeast, Fungi Eukaryotic Multicellular Absorption Eukarya molds Plantae Eukaryotic Multicellular Photosynthesis Plants Animalia Eukaryotic Multicellular Ingestion Animals Prokaryotic Organisms – the archaea, non-photosynthetic bacteria, and cyanobacteria Archaea - Microorganisms that resemble bacteria, but are different from them in certain aspects. Archaea cell walls do not include the macromolecule peptidoglycan, which is always found in the cell walls of bacteria. Archaea usually live in extreme, often very hot or salty environments, such as hot mineral springs or deep-sea hydrothermal vents.