Methicillin-Resistant Staphylococcus Aureus and Clostridium Difficile Infection Lab ID Event Report
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Drug-Resistant Streptococcus Pneumoniae and Methicillin
NEWS & NOTES Conference Summary pneumoniae can vary among popula- conference sessions was that statically tions and is influenced by local pre- sound methods of data collection that Drug-resistant scribing practices and the prevalence capture valid, meaningful, and useful of resistant clones. Conference pre- data and meet the financial restric- Streptococcus senters discussed the role of surveil- tions of state budgets are indicated. pneumoniae and lance in raising awareness of the Active, population-based surveil- Methicillin- resistance problem and in monitoring lance for collecting relevant isolates is the effectiveness of prevention and considered the standard criterion. resistant control programs. National- and state- Unfortunately, this type of surveil- Staphylococcus level epidemiologists discussed the lance is labor-intensive and costly, aureus benefits of including state-level sur- making it an impractical choice for 1 veillance data with appropriate antibi- many states. The challenges of isolate Surveillance otic use programs designed to address collection, packaging and transport, The Centers for Disease Control the antibiotic prescribing practices of data collection, and analysis may and Prevention (CDC) convened a clinicians. The potential for local sur- place an unacceptable workload on conference on March 12–13, 2003, in veillance to provide information on laboratory and epidemiology person- Atlanta, Georgia, to discuss improv- the impact of a new pneumococcal nel. ing state-based surveillance of drug- vaccine for children was also exam- Epidemiologists from several state resistant Streptococcus pneumoniae ined; the vaccine has been shown to health departments that have elected (DRSP) and methicillin-resistant reduce infections caused by resistance to implement enhanced antimicrobial Staphylococcus aureus (MRSA). -

Association Between Carriage of Streptococcus Pneumoniae and Staphylococcus Aureus in Children
BRIEF REPORT Association Between Carriage of Streptococcus pneumoniae and Staphylococcus aureus in Children Gili Regev-Yochay, MD Context Widespread pneumococcal conjugate vaccination may bring about epidemio- Ron Dagan, MD logic changes in upper respiratory tract flora of children. Of particular significance may Meir Raz, MD be an interaction between Streptococcus pneumoniae and Staphylococcus aureus, in view of the recent emergence of community-acquired methicillin-resistant S aureus. Yehuda Carmeli, MD, MPH Objective To examine the prevalence and risk factors of carriage of S pneumoniae Bracha Shainberg, PhD and S aureus in the prevaccination era in young children. Estela Derazne, MSc Design, Setting, and Patients Cross-sectional surveillance study of nasopharyn- geal carriage of S pneumoniae and nasal carriage of S aureus by 790 children aged 40 Galia Rahav, MD months or younger seen at primary care clinics in central Israel during February 2002. Ethan Rubinstein, MD Main Outcome Measures Carriage rates of S pneumoniae (by serotype) and S aureus; risk factors associated with carriage of each pathogen. TREPTOCOCCUS PNEUMONIAE AND Results Among 790 children screened, 43% carried S pneumoniae and 10% car- Staphylococcus aureus are com- ried S aureus. Staphylococcus aureus carriage among S pneumoniae carriers was 6.5% mon inhabitants of the upper vs 12.9% in S pneumoniae noncarriers. Streptococcus pneumoniae carriage among respiratory tract in children and S aureus carriers was 27.5% vs 44.8% in S aureus noncarriers. Only 2.8% carried both Sare responsible for common infec- pathogens concomitantly vs 4.3% expected dual carriage (P=.03). Risk factors for tions. Carriage of S aureus and S pneu- S pneumoniae carriage (attending day care, having young siblings, and age older than moniae can result in bacterial spread and 3 months) were negatively associated with S aureus carriage. -

Clostridium Difficile Infection: How to Deal with the Problem DH INFORMATION RE ADER B OX
Clostridium difficile infection: How to deal with the problem DH INFORMATION RE ADER B OX Policy Estates HR / Workforce Commissioning Management IM & T Planning / Finance Clinical Social Care / Partnership Working Document Purpose Best Practice Guidance Gateway Reference 9833 Title Clostridium difficile infection: How to deal with the problem Author DH and HPA Publication Date December 2008 Target Audience PCT CEs, NHS Trust CEs, SHA CEs, Care Trust CEs, Medical Directors, Directors of PH, Directors of Nursing, PCT PEC Chairs, NHS Trust Board Chairs, Special HA CEs, Directors of Infection Prevention and Control, Infection Control Teams, Health Protection Units, Chief Pharmacists Circulation List Description This guidance outlines newer evidence and approaches to delivering good infection control and environmental hygiene. It updates the 1994 guidance and takes into account a national framework for clinical governance which did not exist in 1994. Cross Ref N/A Superseded Docs Clostridium difficile Infection Prevention and Management (1994) Action Required CEs to consider with DIPCs and other colleagues Timing N/A Contact Details Healthcare Associated Infection and Antimicrobial Resistance Department of Health Room 528, Wellington House 133-155 Waterloo Road London SE1 8UG For Recipient's Use Front cover image: Clostridium difficile attached to intestinal cells. Reproduced courtesy of Dr Jan Hobot, Cardiff University School of Medicine. Clostridium difficile infection: How to deal with the problem Contents Foreword 1 Scope and purpose 2 Introduction 3 Why did CDI increase? 4 Approach to compiling the guidance 6 What is new in this guidance? 7 Core Guidance Key recommendations 9 Grading of recommendations 11 Summary of healthcare recommendations 12 1. -

Methicillin-Resistant Staphylococcus Aureus (MRSA)
Methicillin-Resistant Staphylococcus Aureus (MRSA) Over the past several decades, the incidence of resistant gram-positive organisms has risen in the United States. MRSA strains, first identified in the 1960s in England, were first observed in the U.S. in the mid 1980s.1 Resistance quickly developed, increasing from 2.4% in 1979 to 29% in 1991.2 The current prevalence for MRSA in hospitals and other facilities ranges from <10% to 65%. In 1999, MRSA accounted for more than 50% of all Staphylococcus aureus isolates within U.S. intensive care units.3, 4 The past years, however, outbreaks of MRSA have also been seen in the community setting, particularly among preschool-age children, some of whom have attended day-care centers.5, 6, 7 MRSA does not appear to be more virulent than methicillin-sensitive Staphylococcus aureus, but certainly poses a greater treatment challenge. MRSA also has been associated with higher hospital costs and mortality.8 Within a decade of its development, methicillin resistance to Staphylococcus aureus emerged.9 MRSA strains generally are now resistant to other antimicrobial classes including aminoglycosides, beta-lactams, carbapenems, cephalosporins, fluoroquinolones and macrolides.10,11 Most of the resistance was secondary to production of beta-lactamase enzymes or intrinsic resistance with alterations in penicillin-binding proteins. Staphylococcus aureus is the most frequent cause of nosocomial pneumonia and surgical- wound infections and the second most common cause of nosocomial bloodstream infections.12 Long-term care facilities (LTCFs) have developed rates of MRSA ranging from 25%-35%. MRSA rates may be higher in LTCFs if they are associated with hospitals that have higher rates.13 Transmission of MRSA generally occurs through direct or indirect contact with a reservoir. -

Methicillin Resistant Staphylococcus Aureus (MRSA)
Methicillin resistant Staphylococcus aureus (MRSA), Clostridium difficile and ESBL-producing Escherichia coli in the home and community: assessing the problem, controlling the spread An expert report commissioned by the International Scientific Forum on Home Hygiene © International Scientific Forum on Home Hygiene, 2006 2 CONTENTS ACKNOWLEDGMENTS 4 SUMMARY 5 1. INTRODUCTION – INFECTIOUS DISEASE AND HYGIENE IN THE 21ST CENTURY 7 2. THE CHAIN OF INFECTION TRANSMISSION IN THE HOME 10 3. METHICILLIN RESISTANT STAPHYLOCOCCUS AUREUS (MRSA) 11 3.1 CHARACTERISTICS OF STAPHYLOCOCCUS AUREUS AND METHICILLIN RESISTANT STAPHYLOCOCCUS AUREUS 11 3.2 HOSPITAL-ACQUIRED, HEALTHCARE ASSOCIATED AND COMMUNITY ACQUIRED MRSA 11 3.3 UNDERSTANDING THE CHAIN OF INFECTION FOR S. AUREUS AND MRSA IN THE HOME 13 3.4 WHAT ARE THE RISKS ASSOCIATED WITH S. AUREUS AND MRSA IN THE COMMUNITY? 22 4. CLOSTRIDIUM DIFFICILE 24 4.1 CHARACTERISTICS OF CLOSTRIDIUM DIFFICILE 24 4.2 HOSPITAL-ACQUIRED AND COMMUNITY- ACQUIRED C. DIFFICILE 24 4.3 UNDERSTANDING THE CHAIN OF INFECTION FOR C. DIFFICILE IN THE HOME 25 4.4 WHAT ARE THE RISKS ASSOCIATED WITH C. DIFFICILE IN THE HOME AND COMMUNITY 28 5. EXTENDED-SPECTRUM ß-LACTAMASE ESCHERICHIA COLI (ESBLS) 30 5.1 CHARACTERISTICS OF ESCHERICHIA COLI 30 5.2 HOSPITAL AND COMMUNITY-ACQUIRED ESBL-PRODUCING E. COLI INFECTIONS 31 5.3 UNDERSTANDING THE CHAIN OF INFECTION FOR E. COLI AND ESBL-PRODUCING E. COLI IN THE HOME 31 5.4 RISKS ASSOCIATED WITH ESBLS IN THE COMMUNITY 33 6. REDUCING THE RISKS OF TRANSMISSION OF METHICILLIN RESISTANT STAPHYLOCOCCUS AUREUS (MRSA), CLOSTRIDIUM DIFFICILE AND ESBL-PRODUCING ESCHERICHIA COLI IN THE HOME AND COMMUNITY - DEVELOPING A RISK-BASED APPROACH TO HOME HYGIENE 35 6.1 BREAKING THE CHAIN OF TRANSMISSION OF INFECTION IN THE HOME 35 6.2 DEVELOPING A RISK-BASED APPROACH TO HOME HYGIENE 35 6.3 INTERRUPTING THE CHAIN OF INFECTION TRANSMISSION IN THE HOME 38 7. -
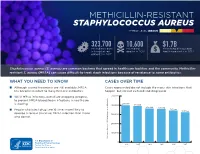
Methicillin-Resistant Staphylococcus Aureus Threat Level Serious
METHICILLIN-RESISTANT STAPHYLOCOCCUS AUREUS THREAT LEVEL SERIOUS 323,700 10,600 $1.7B Estimated cases Estimated Estimated attributable in hospitalized deaths in 2017 healthcare costs in 2017 patients in 2017 Staphylococcus aureus (S. aureus) are common bacteria that spread in healthcare facilities and the community. Methicillin- resistant S. aureus (MRSA) can cause difficult-to-treat staph infections because of resistance to some antibiotics. WHAT YOU NEED TO KNOW CASES OVER TIME ■ Although several treatments are still available, MRSA Cases represented do not include the many skin infections that has become resistant to many first-line antibiotics. happen, but are not cultured and diagnosed. ■ While MRSA infections overall are dropping, progress 500,000 to prevent MRSA bloodstream infections in healthcare 401,000 is slowing. 400,000 391,000 365,400 359,500 ■ People who inject drugs are 16 times more likely to 343,100 323,700 develop a serious (invasive) MRSA infection than those 300,000 who do not. 200,000 100,000 Estimated Cases of MRSA in Hospitalized Patients in Hospitalized Cases of MRSA Estimated 0 2012 2013 2014 2015 2016 2017 30 25 An estimated 7% decline each year 20 15 10 An estimated 17% decline each year from 2005 to 2016, and no change from 2013 to 2016 5 Estimated Cases of MRSA Per 100,000 People 100,000 Per Cases of MRSA Estimated 0 2005 2006 2007 2008 2009 2010 2011 2012 2013 2014 2015 2016 Community-onset MRSA Bloodstream Infection Rate Hospital-onset MRSA Bloodstream Infection Rate 500,000 401,000 400,000 391,000 365,400 359,500 343,100 323,700 300,000 METHICILLIN-RESISTANT STAPHYLOCOCCUS AUREUS200,000 (MRSA) MRSA INFECTIONS CAN BE PREVENTED REDUCTIONS100,000 IN HOSPITALS HAVE STALLED MRSA infections are preventable and many lives have been New Patients in Hospitalized Cases of MRSA Estimated strategies in healthcare, along with current CDC 0 saved through effective infection control interventions. -

Antibiotic Resistance Threats in the United States, 2019
ANTIBIOTIC RESISTANCE THREATS IN THE UNITED STATES 2019 Revised Dec. 2019 This report is dedicated to the 48,700 families who lose a loved one each year to antibiotic resistance or Clostridioides difficile, and the countless healthcare providers, public health experts, innovators, and others who are fighting back with everything they have. Antibiotic Resistance Threats in the United States, 2019 (2019 AR Threats Report) is a publication of the Antibiotic Resistance Coordination and Strategy Unit within the Division of Healthcare Quality Promotion, National Center for Emerging and Zoonotic Infectious Diseases, Centers for Disease Control and Prevention. Suggested citation: CDC. Antibiotic Resistance Threats in the United States, 2019. Atlanta, GA: U.S. Department of Health and Human Services, CDC; 2019. Available online: The full 2019 AR Threats Report, including methods and appendices, is available online at www.cdc.gov/DrugResistance/Biggest-Threats.html. DOI: http://dx.doi.org/10.15620/cdc:82532. ii U.S. Centers for Disease Control and Prevention Contents FOREWORD .............................................................................................................................................V EXECUTIVE SUMMARY ........................................................................................................................ VII SECTION 1: THE THREAT OF ANTIBIOTIC RESISTANCE ....................................................................1 Introduction .................................................................................................................................................................3 -

Detection of Causative Agents in Focus in Erysipelas Patients By
Detection of causative agents out using the ATB Expression ( «BioMerieux») Conclusion. The primary role of the β-HSA DNA contamination of hearth of erysipelas is and test systems ID32E, ID32Staph, ID32Strep, is indisputable, in the cases with complications depends on the presence of live staphylococci in focus in erysipelas patients ID32GN, identification of β-HS in groups A, B, C, mixt-infection can take a part in the etiology of on skin surface by bacteriological method and D, F and G (at Lancefield) - a latex agglutination the disease. PCR-RT with fluorescent Staphylococci on skin: PCR reaction(Pastorex Strep Kits; «Bio-Rad»,France) hybridization detection showed the highly informative content in the study of the blood and S.aureus No staphyl. S.epid./saprop. Volchkova E.V.1, Belaia O.F.1, Troickii V.I.1, The overall frequency of detection of different Svistunova Т.S.2,Potekaeva S.N.2, microorganisms (85%) in PCR in patients bulls punctuate. and it was more informative in Staphylococcus DNA in hearth: Domonova E.A.3 with erysipelas (n=60) identifying the microflora in patients with 1 erysipelas than bacteriological method. The 80% 32% # 7,0% # Sechenov First Moscow State Medical Staphylococci S.aureus-39% University, 2 Infectious Clinical Hospital №2, bacteriological method showed low efficacy in 74.5% MSSA:MRSA = 9:2 PCR results: 3Central Research Institute of Epidemiology, detecting of Str.pyogenes, however, positive S.epidermidis and/or Moscow, Russian Federation results of bacteriological research can serve as MRSA -2 - - S.saprophyticus-43% MRCoNS - 4 - MRCoNS -5 Erysipelas is a common infection of the skin an objective criterion of therapy correction based MRCoNS-20% MSSA - 7 MSSA -1 MSSA - 2 surface layer, unlike cellulitis and necrotizing on identified microbes. -

Pneumonia Panel
Guidance on Use of the Pneumonia Panel for Respiratory Infections Although the number of pathogens that cause pneumonia is lengthy, establishing the microbiologic etiology of pneumonia is inherently difficult. A recent large multi-center study of community-acquired pneumonia (CAP) found that only 38% of 2259 CAP cases had a microbiologic diagnosis with 23% having viruses detected, 11% bacterial, and 3% had both viruses and bacteria detected.1 Current tools to assist in pneumonia diagnosis include respiratory tract cultures (sputum, BAL, tracheal aspirate, mini-BAL), urine antigens (pneumococcal, Legionella), serology, and PCR for viral and certain bacterial pathogens. While these tools are useful, the study noted above used all these tools and was unable to document an etiology causing pneumonia in 62% of patients. Thus, more sensitive tools for detection of respiratory pathogens are still needed. Nebraska Medicine has recently introduced a new FDA-approved multiplex PCR panel to assist in determination of the etiology of pneumonia, termed the Pneumonia Panel (PP). This test uses a nested multiplex PCR- approach to amplify nucleic acid targets directly from sputum or bronchoalveolar lavage (BAL) in patients with suspected pneumonia. The list of pathogens and resistance genes included in the panel is found in Table 1. Note that the bacterial targets are detected semi-quantitatively whereas the atypical pathogens and the viral targets are detected qualitatively. Table 1: Pneumonia Panel Pathogen Targets and Associated Resistance Genes Semi-quantitative Detection: Gram Positive Organisms: Resistance Genes (Staph aureus only): Staphylococcus aureus mecA/C and MREJ Streptococcus pneumoniae Streptococcus agalactiae Streptococcus pyogenes Gram Negative Organisms: Resistance Genes (All Gram Negatives): Acinetobacter calcoaceticus-baumannii complex CTX-M Enterobacter cloacae complex IMP E. -

Methodology for Staphylococcus Areus and Clostridium Perfringens
342 MFTHODOLQGY FOR STAPHYLOCOCCUS AU€WJS AND CUIZX'RIDIUM PERF'RINGE:NSK H. W. WALKER AND C. R. EU3Y Iowa State University The staphylococci and micrococcihave been-placed in the family Micrococcaceae in "Bergey ' s Manual of Determinative Bacteriology" (Breed et g., 1957). Classification of these gram-positive, catalase-positive cocci in this family is still somewhat controversial. In 1965, however, the International Subcommittee on Staphylococci and Micrococci proposed a standard test for separating these organisms into the twcJ genera :,f Staphylococcus and Micrococcus. According to this cormnittee, the genus Staphylococcus should contain the parasitic, facultative-anaerobic cocci which produce acid from glucose under anaerobic conditions, and the genus Micrococcus should contain the saprophytic, aerobic cocci which produce acid from glucose aerobically, but not anaerobically. Our interest is mainly with the genus Sta h lococcus which consists of two well accepted species, S. aureus and S. -$=-7-e idermidis Breed et al., 1957). These tw:, species are separgtted as follows: S. aureus ferments mannitol and is coagulase positive, while S. epidermidis does not ferment mannitol and is coagulase negative. S . aureug produces a variety of extracellular products, among which is enterotoxin, a heat-stable exotoxin that causes a severe type of foQd poisoning in man and ~omeother animal species. Animal assays are not convenient for testing for strains that produce this toxin; therefore, other simplified means of detecting pathogenic strains have been used or suggested. Coagulase production is probably the most extensively used test for correlation with enterotoxigenicity (Chapman, 1944; Morrison, 1962); this enzyme causes the formation of a clot or coagulum in citrated or oxalated blood plasma. -

Guidelines for Prevention and Control of Infection Due to Antibiotic Resistant
Guidelines for Prevention and Control of Infection Due to Antibiotic Resistant Organisms Updated December 2020 Florida Department of Health Division of Disease Control & Health Protection Bureau of Epidemiology 850-245-4401 (24/7) FLHealth.gov Table of Contents I. Statement of Purpose ................................................................................................... 1 II. Definitions ................................................................................................................... 2 III. Introduction ................................................................................................................ 8 IV. Background ................................................................................................................ 9 A. Methicillin•Resistant Staphylococcus aureus (MRSA) .......................................... 9 B. Vancomycin•Resistant Enterococci (VRE) ......................................................... 10 C. Clostridioides difficile .......................................................................................... 10 D. Multidrug•Resistant Gram•Negative Bacilli (MDR•GNB) .................................... 11 1. Acinetobacter baumannii ........................................................................... 11 2. Carbapenem•Resistant Enterobacteriaceae (CRE) ................................... 12 E. Candida auris (C. auris) ..................................................................................... 13 V. Colonization vs. Infection ......................................................................................... -

Staphylococcus Aureus Toxins and Their Molecular Activity in Infectious Diseases
toxins Review Staphylococcus aureus Toxins and Their Molecular Activity in Infectious Diseases Diana Oliveira ID , Anabela Borges ID and Manuel Simões * ID LEPABE, Department of Chemical Engineering, Faculty of Engineering, University of Porto, Rua Dr. Roberto Frias, s/n, 4200-465 Porto, Portugal; [email protected] (D.O.); [email protected] (A.B.) * Correspondence: [email protected] Received: 30 April 2018; Accepted: 15 June 2018; Published: 19 June 2018 Abstract: Staphylococcus aureus is a microorganism resident in the skin and nasal membranes with a dreadful pathogenic potential to cause a variety of community and hospital-acquired infections. The frequency of these infections is increasing and their treatment is becoming more difficult. The ability of S. aureus to form biofilms and the emergence of multidrug-resistant strains are the main reasons determining the challenge in dealing with these infections. S. aureus' infectious capacity and its success as a pathogen is related to the expression of virulence factors, among which the production of a wide variety of toxins is highlighted. For this reason, a better understanding of S. aureus toxins is needed to enable the development of new strategies to reduce their production and consequently improve therapeutic approaches. This review focuses on understanding the toxin-based pathogenesis of S. aureus and their role on infectious diseases. Keywords: biofilms; epidemiology; exfoliative toxins; pore-forming toxins; Staphylococcus aureus; superantigens Key Contribution: This review analyzes the mechanisms of toxin production by S. aureus and describes their molecular activity in infectious diseases. 1. Introduction S. aureus is an opportunistic human pathogen known to colonize the respiratory tract of approximately 30% of the U.S.