Alternative Splicing Events Expand Molecular Diversity of Camel
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Bison Literature Review Biology
Bison Literature Review Ben Baldwin and Kody Menghini The purpose of this document is to compare the biology, ecology and basic behavior of cattle and bison for a management context. The literature related to bison is extensive and broad in scope covering the full continuum of domestication. The information incorporated in this review is focused on bison in more or less “wild” or free-ranging situations i.e.., not bison in close confinement or commercial production. While the scientific literature provides a solid basis for much of the basic biology and ecology, there is a wealth of information related to management implications and guidelines that is not captured. Much of the current information related to bison management, behavior (especially social organization) and practical knowledge is available through local experts, current research that has yet to be published, or popular literature. These sources, while harder to find and usually more localized in scope, provide crucial information pertaining to bison management. Biology Diet Composition Bison evolutional history provides the basis for many of the differences between bison and cattle. Bison due to their evolution in North America ecosystems are better adapted than introduced cattle, especially in grass dominated systems such as prairies. Many of these areas historically had relatively low quality forage. Bison are capable of more efficient digestion of low-quality forage then cattle (Peden et al. 1973; Plumb and Dodd 1993). Peden et al. (1973) also found that bison could consume greater quantities of low protein and poor quality forage then cattle. Bison and cattle have significant dietary overlap, but there are slight differences as well. -

Last Interglacial (MIS 5) Ungulate Assemblage from the Central Iberian Peninsula: the Camino Cave (Pinilla Del Valle, Madrid, Spain)
Palaeogeography, Palaeoclimatology, Palaeoecology 374 (2013) 327–337 Contents lists available at SciVerse ScienceDirect Palaeogeography, Palaeoclimatology, Palaeoecology journal homepage: www.elsevier.com/locate/palaeo Last Interglacial (MIS 5) ungulate assemblage from the Central Iberian Peninsula: The Camino Cave (Pinilla del Valle, Madrid, Spain) Diego J. Álvarez-Lao a,⁎, Juan L. Arsuaga b,c, Enrique Baquedano d, Alfredo Pérez-González e a Área de Paleontología, Departamento de Geología, Universidad de Oviedo, C/Jesús Arias de Velasco, s/n, 33005 Oviedo, Spain b Centro Mixto UCM-ISCIII de Evolución y Comportamiento Humanos, C/Sinesio Delgado, 4, 28029 Madrid, Spain c Departamento de Paleontología, Facultad de Ciencias Geológicas, Universidad Complutense de Madrid, Ciudad Universitaria, 28040 Madrid, Spain d Museo Arqueológico Regional de la Comunidad de Madrid, Plaza de las Bernardas, s/n, 28801-Alcalá de Henares, Madrid, Spain e Centro Nacional de Investigación sobre la Evolución Humana (CENIEH), Paseo Sierra de Atapuerca, s/n, 09002 Burgos, Spain article info abstract Article history: The fossil assemblage from the Camino Cave, corresponding to the late MIS 5, constitutes a key record to un- Received 2 November 2012 derstand the faunal composition of Central Iberia during the last Interglacial. Moreover, the largest Iberian Received in revised form 21 January 2013 fallow deer fossil population was recovered here. Other ungulate species present at this assemblage include Accepted 31 January 2013 red deer, roe deer, aurochs, chamois, wild boar, horse and steppe rhinoceros; carnivores and Neanderthals Available online 13 February 2013 are also present. The origin of the accumulation has been interpreted as a hyena den. Abundant fallow deer skeletal elements allowed to statistically compare the Camino Cave fossils with other Keywords: Early Late Pleistocene Pleistocene and Holocene European populations. -
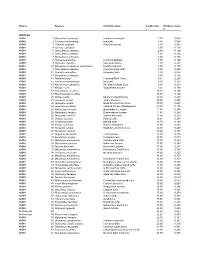
PDF File Containing Table of Lengths and Thicknesses of Turtle Shells And
Source Species Common name length (cm) thickness (cm) L t TURTLES AMNH 1 Sternotherus odoratus common musk turtle 2.30 0.089 AMNH 2 Clemmys muhlenbergi bug turtle 3.80 0.069 AMNH 3 Chersina angulata Angulate tortoise 3.90 0.050 AMNH 4 Testudo carbonera 6.97 0.130 AMNH 5 Sternotherus oderatus 6.99 0.160 AMNH 6 Sternotherus oderatus 7.00 0.165 AMNH 7 Sternotherus oderatus 7.00 0.165 AMNH 8 Homopus areolatus Common padloper 7.95 0.100 AMNH 9 Homopus signatus Speckled tortoise 7.98 0.231 AMNH 10 Kinosternon subrabum steinochneri Florida mud turtle 8.90 0.178 AMNH 11 Sternotherus oderatus Common musk turtle 8.98 0.290 AMNH 12 Chelydra serpentina Snapping turtle 8.98 0.076 AMNH 13 Sternotherus oderatus 9.00 0.168 AMNH 14 Hardella thurgi Crowned River Turtle 9.04 0.263 AMNH 15 Clemmys muhlenbergii Bog turtle 9.09 0.231 AMNH 16 Kinosternon subrubrum The Eastern Mud Turtle 9.10 0.253 AMNH 17 Kinixys crosa hinged-back tortoise 9.34 0.160 AMNH 18 Peamobates oculifers 10.17 0.140 AMNH 19 Peammobates oculifera 10.27 0.140 AMNH 20 Kinixys spekii Speke's hinged tortoise 10.30 0.201 AMNH 21 Terrapene ornata ornate box turtle 10.30 0.406 AMNH 22 Terrapene ornata North American box turtle 10.76 0.257 AMNH 23 Geochelone radiata radiated tortoise (Madagascar) 10.80 0.155 AMNH 24 Malaclemys terrapin diamondback terrapin 11.40 0.295 AMNH 25 Malaclemys terrapin Diamondback terrapin 11.58 0.264 AMNH 26 Terrapene carolina eastern box turtle 11.80 0.259 AMNH 27 Chrysemys picta Painted turtle 12.21 0.267 AMNH 28 Chrysemys picta painted turtle 12.70 0.168 AMNH 29 -

European Bison
IUCN/Species Survival Commission Status Survey and Conservation Action Plan The Species Survival Commission (SSC) is one of six volunteer commissions of IUCN – The World Conservation Union, a union of sovereign states, government agencies and non- governmental organisations. IUCN has three basic conservation objectives: to secure the conservation of nature, and especially of biological diversity, as an essential foundation for the future; to ensure that where the Earth’s natural resources are used this is done in a wise, European Bison equitable and sustainable way; and to guide the development of human communities towards ways of life that are both of good quality and in enduring harmony with other components of the biosphere. A volunteer network comprised of some 8,000 scientists, field researchers, government officials Edited by Zdzis³aw Pucek and conservation leaders from nearly every country of the world, the SSC membership is an Compiled by Zdzis³aw Pucek, Irina P. Belousova, unmatched source of information about biological diversity and its conservation. As such, SSC Ma³gorzata Krasiñska, Zbigniew A. Krasiñski and Wanda Olech members provide technical and scientific counsel for conservation projects throughout the world and serve as resources to governments, international conventions and conservation organisations. IUCN/SSC Action Plans assess the conservation status of species and their habitats, and specifies conservation priorities. The series is one of the world’s most authoritative sources of species conservation information -

Phylogeny of Dictyocaulus (Lungworms) from Eight Species of Ruminants Based on Analyses of Ribosomal RNA Data
179 Phylogeny of Dictyocaulus (lungworms) from eight species of ruminants based on analyses of ribosomal RNA data J. HO¨ GLUND1*,D.A.MORRISON1, B. P. DIVINA1,2, E. WILHELMSSON1 and J. G. MATTSSON1 1 Department of Parasitology (SWEPAR), National Veterinary Institute and Swedish University of Agriculture Sciences, 751 89 Uppsala, Sweden 2 College of Veterinary Medicine, University of the Philippines Los Ban˜os, College, Laguna, 4031 Philippines (Received 8 November 2002; revised 8 February 2003; accepted 8 February 2003) SUMMARY In this study, we conducted phylogenetic analyses of nematode parasites within the genus Dictyocaulus (superfamily Trichostrongyloidea). Lungworms from cattle (Bos taurus), domestic sheep (Ovis aries), European fallow deer (Dama dama), moose (Alces alces), musk ox (Ovibos moschatus), red deer (Cervus elaphus), reindeer (Rangifer tarandus) and roe deer (Capreolus capreolus) were obtained and their small subunit ribosomal RNA (SSU) and internal transcribed spacer 2 (ITS2) sequences analysed. In the hosts examined we identified D. capreolus, D. eckerti, D. filaria and D. viviparus. However, in fallow deer we detected a taxon with unique SSU and ITS2 sequences. The phylogenetic position of this taxon based on the SSU sequences shows that it is a separate evolutionary lineage from the other recognized species of Dictyocaulus. Furthermore, the analysis of the ITS2 sequence data indicates that it is as genetically distinct as are the named species of Dictyocaulus. Therefore, either this taxon needs to be recognized as a new species, or D. capreolus, D. eckerti and D. viviparus need to be combined into a single species. Traditionally, the genus Dictyocaulus has been placed as a separate family within the superfamily Trichostrongyloidea. -

Mixed-Species Exhibits with Pigs (Suidae)
Mixed-species exhibits with Pigs (Suidae) Written by KRISZTIÁN SVÁBIK Team Leader, Toni’s Zoo, Rothenburg, Luzern, Switzerland Email: [email protected] 9th May 2021 Cover photo © Krisztián Svábik Mixed-species exhibits with Pigs (Suidae) 1 CONTENTS INTRODUCTION ........................................................................................................... 3 Use of space and enclosure furnishings ................................................................... 3 Feeding ..................................................................................................................... 3 Breeding ................................................................................................................... 4 Choice of species and individuals ............................................................................ 4 List of mixed-species exhibits involving Suids ........................................................ 5 LIST OF SPECIES COMBINATIONS – SUIDAE .......................................................... 6 Sulawesi Babirusa, Babyrousa celebensis ...............................................................7 Common Warthog, Phacochoerus africanus ......................................................... 8 Giant Forest Hog, Hylochoerus meinertzhageni ..................................................10 Bushpig, Potamochoerus larvatus ........................................................................ 11 Red River Hog, Potamochoerus porcus ............................................................... -

Idiograms of Yak (Bos Grunniens), Cattle (Bos Taurus) and Their Hybrid C.P
IDIOGRAMS OF YAK (BOS GRUNNIENS), CATTLE (BOS TAURUS) AND THEIR HYBRID C.P. Popescu To cite this version: C.P. Popescu. IDIOGRAMS OF YAK (BOS GRUNNIENS), CATTLE (BOS TAURUS) AND THEIR HYBRID. Annales de génétique et de sélection animale, INRA Editions, 1969, 1 (3), pp.207-217. hal- 00892361 HAL Id: hal-00892361 https://hal.archives-ouvertes.fr/hal-00892361 Submitted on 1 Jan 1969 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. IDIOGRAMS OF YAK (BOS GRUNNIENS), CATTLE (BOS TAURUS) AND THEIR HYBRID C.P. POPESCU Department of Genetics, Institute of Zootechnical Researches, 63, Dr Staicovici, Sector 6-Bucarest (Romania) (!‘) ABSTRACT The karyotypes of Yak (Bos grunniens L.) and Cattle (Bos taurus L.) are alike both numeri- cally and morphologically. However, idiograms of the two species reveal differences both in autosomes and in sexual chromosomes. The idiogram of hybrids (Bos grunniens L. S x Bos taurus L. !) represents roughly an average of parental idiograms. Numerous authors have studied the chromosomes of some hybrid animals and their parental species for the purpose of explaining the infertility of inter- specific hybrids (BASRUR and MooN, 1967; LAY and NADr,!R, ig6g; MA!NO et al., 1963). -

Holocene Distribution of European Bison – the Archaeozoological Record
MUNIBE (Antropologia-Arkeologia) 57 Homenaje a Jesús Altuna 421-428 SAN SEBASTIAN 2005 ISSN 1132-2217 The Holocene distribution of European bison – the archaeozoological record Distribución Holocena del bisonte europeo - el registro arqueozoológico KEY WORDS: Europe, Holocene, European bison, distribution, archaeozoological record. PALABRAS CLAVE: Europa, Holoceno, bisonte europeo, distribución, registro arqueozoológico. Norbert BENECKE* ABSTRACT The paper presents a reconstruction of the Holocene distribution of European bison or wisent. It is based on the archaeozoological record of this species. European bison was an early Postglacial immigrant into the European continent. The oldest evidence comes from sites in northern Central Europe and South Scandinavia dating to the Preboreal. In the Mid- and Late Holocene, European bison was widely distributed on the European continent. Its range extended from France in the west to the Ukraine and Russia in the east. Except for an area comprising East Poland, Belarus, Lithuania and Latvia, European bison was a rare species in most regions of its range. In the Middle Ages, there is a shrinkage of the range of wisent in its western part. RESUMEN El artículo presenta una reconstrucción de la distribución holocena del bisonte europeo. Está basada en el registro arqueozoológico de es- ta especie. El bisonte europeo fue un inmigrante al Continente europeo durante el Postglacial inicial. La más antigua evidencia procede de ya- cimientos del Norte de Europa Central y del Sur de Escandinavia, que datan del Preboreal. Durante el Holoceno medio y tardío el bisonte eu- ropeo estaba ampliamente distribuido en el Continente europeo. Su distribución se extendía desde Francia al W hasta Ucrania al E. -

Spatial Habitat Overlap & Habitat Preference of Himalayan Musk Deer
Current Research Journal of Biological Sciences 2(3): 217-225, 2010 ISSN: 2041-0778 © Maxwell Scientific Organization, 2010 Submitted Date: April 23, 2010 Accepted Date: May 08, 2010 Published Date: May 20, 2010 Spatial Habitat Overlap and Habitat Preference of Himalayan Musk Deer (Moschus chrysogaster) in Sagarmatha (Mt. Everest) National Park, Nepal 1,2Achyut Aryal, 3David Raubenheimer, 4Suman Subedi and 5Bijaya Kattel 1Ecology and Conservation Group, Institute of Natural Sciences, Massey University, New Zealand 2The Biodiversity Research and Training Forum (BRTF), Nepal 3Nutritional Ecology Research Group, Institute of Natural Sciences, Massey University, New Zealand 4 Sagarmatha (Mt. Everest) National Park, Department of National Park and Wildlife Conservation, Government of Nepal 5South Florida Water Management District West Palm Beach, Florida, USA Abstract: The musk deer (Moschus chrysogaster), which is native to Nepal, China, Bhutan, and India, is an endangered species, which suffers a high level of poaching due to the economic demand for its musk pod. The World Heritage Site (WHS), Sagarmatha (Mt. Everest) National Park (SNP), provides prime habitat for this species. Our aim in this study was to perform a quantitative assessment of the habitat preferences of musk deer in SNP, and evaluate how preferred habitat might be impacted by anthropogenic activities. Results showed that the musk deer population is distributed in 131 km2 of the park area. We recorded 39 musk deer (11 male, 16 female and 12 unidentified) in Debuche, Tengboche, Phortse Thanga, Dole, and associated areas in SNP. The musk deer in these areas preferred gentle to steep slopes with the altitudinal range of 3400-3900m and also displayed a preference for dense forest and sparse ground/crown cover. -

Host Specificity of Enterocytozoon Bieneusi Genotypes in Bactrian
Qi et al. Parasites & Vectors (2018) 11:219 https://doi.org/10.1186/s13071-018-2793-9 RESEARCH Open Access Host specificity of Enterocytozoon bieneusi genotypes in Bactrian camels (Camelus bactrianus)inChina Meng Qi1,2†, Junqiang Li2†, Aiyun Zhao1, Zhaohui Cui2, Zilin Wei1, Bo Jing1* and Longxian Zhang2* Abstract Background: Enterocytozoon bieneusi is an obligate, intracellular fungus and is commonly reported in humans and animals. To date, there have been no reports of E. bieneusi infections in Bactrian camels (Camelus bactrianus). The present study was conducted to understand the occurrence and molecular characteristics of E. bieneusi in Bactrian camels in China. Results: Of 407 individual Bactrian camel fecal specimens, 30.0% (122) were E. bieneusi-positive by nested polymerase chain reaction (PCR) based on internal transcriber spacer (ITS) sequence analysis. A total of 14 distinct E. bieneusi ITS genotypes were obtained: eight known genotypes (genotype EbpC, EbpA, Henan-IV, BEB6, CM8, CHG16, O and WL17), and six novel genotypes (named CAM1 to CAM6). Genotype CAM1 (59.0%, 72/122) was the most predominant genotype in Bactrian camels in Xinjiang, and genotype EbpC (18.9%, 23/122) was the second-most predominant genotype. Phylogenetic analysis revealed that six known genotypes (EbpC, EbpA, WL17, Henan-IV, CM8 and O) and three novel genotypes (CAM3, CAM5 and CAM6) fell into the human-pathogenic group 1. Two known genotypes (CHG16 and BEB6) fell into the cattle host-specific group 2. The novel genotypes CAM1, CAM 2 and CAM4 cluster into group 8. Conclusions: To our knowledge, this is the first report of E. bieneusi in Bactrian camels. -

Gaur (<I>Bos Gaurus</I>) Abundance, Distribution, and Habitat Use
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Dissertations & Theses in Natural Resources Natural Resources, School of 5-2016 Gaur (Bos gaurus) Abundance, Distribution, and Habitat use patterns in Kuiburi National Park, Southwestern Thailand Supatcharee Tanasarnpaiboon University of Nebraska-Lincoln, [email protected] Follow this and additional works at: http://digitalcommons.unl.edu/natresdiss Part of the Environmental Indicators and Impact Assessment Commons, Environmental Monitoring Commons, Natural Resources and Conservation Commons, Natural Resources Management and Policy Commons, and the Other Ecology and Evolutionary Biology Commons Tanasarnpaiboon, Supatcharee, "Gaur (Bos gaurus) Abundance, Distribution, and Habitat use patterns in Kuiburi National Park, Southwestern Thailand" (2016). Dissertations & Theses in Natural Resources. 129. http://digitalcommons.unl.edu/natresdiss/129 This Article is brought to you for free and open access by the Natural Resources, School of at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Dissertations & Theses in Natural Resources by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. GAUR (BOS GAURUS) ABUNDANCE, DISTRIBUTION, AND HABITAT USE PATTERNS IN KUIBURI NATIONAL PARK, SOUTHWESTERN THAILAND by Supatcharee Tanasarnpaiboon A DISSERTATION Presented to the Faculty of The Graduate College at the University of Nebraska In Partial Fulfillment of Requirements For the Degree of Doctor of Philosophy Major: Natural Resource Sciences (Applied Ecology) Under the Supervision of Professor John P. Carroll Lincoln, Nebraska May, 2016 GAUR (Bos gaurus) ABUNDANCE, DISTRIBUTION, AND HABITAT USE PATTERNS IN KUIBURI NATIONAL PARK, SOUTHWESTERN THAILAND Supatcharee Tanasarnpaiboon, Ph.D. University of Nebraska, 2016 Advisor: John P. Carroll Population status of gaur (Bos gaurus), a wild cattle, in most habitats where they are present, is still unknown. -

Antelopes, Gazelles, Cattle, Goats, Sheep, and Relatives
© Copyright, Princeton University Press. No part of this book may be distributed, posted, or reproduced in any form by digital or mechanical means without prior written permission of the publisher. INTRODUCTION RECOGNITION The family Bovidae, which includes Antelopes, Cattle, Duikers, Gazelles, Goats, and Sheep, is the largest family within Artiodactyla and the most diverse family of ungulates, with more than 270 recent species. Their common characteristic is their unbranched, non-deciduous horns. Bovids are primarily Old World in their distribution, although a few species are found in North America. The name antelope is often used to describe many members of this family, but it is not a definable, taxonomically based term. Shape, size, and color: Bovids encompass an extremely wide size range, from the minuscule Royal Antelope and the Dik-diks, weighing as little as 2 kg and standing 25 to 35 cm at the shoulder, to the Asian Wild Water Buffalo, which weighs as much as 1,200 kg, and the Gaur, which measures up to 220 cm at the shoulder. Body shape varies from relatively small, slender-limbed, and thin-necked species such as the Gazelles to the massive, stocky wild cattle (fig. 1). The forequarters may be larger than the hind, or the reverse, as in smaller species inhabiting dense tropical forests (e.g., Duikers). There is also a great variety in body coloration, although most species are some shade of brown. It can consist of a solid shade, or a patterned pelage. Antelopes that rely on concealment to avoid predators are cryptically colored. The stripes and blotches seen on the hides of Bushbuck, Bongo, and Kudu also function as camouflage by helping to disrupt the animals’ outline.