PCR-RFLP Identification of Meat from Red Deer, Sika Deer, Roe Deer, Fallow Deer, Mouflon, Wild Boar, Hare and Cattle
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

BIGHORN SHEEP Ovis Canadensis Original1 Prepared by R.A
BIGHORN SHEEP Ovis canadensis Original1 prepared by R.A. Demarchi Species Information Distribution Global Taxonomy The genus Ovis is present in west-central Asia, Until recently, three species of Bighorn Sheep were Siberia, and North America (and widely introduced recognized in North America: California Bighorn in Europe). Approximately 38 000 Rocky Mountain Sheep (Ovis canadensis californiana), Rocky Bighorn Sheep (Wishart 1999) are distributed in Mountain Bighorn Sheep (O. canadensis canadensis), scattered patches along the Rocky Mountains of and Desert Bighorn Sheep (O. canadensis nelsoni). As North America from west of Grand Cache, Alberta, a result of morphometric measurements, and to northern New Mexico. They are more abundant protein and mtDNA analysis, Ramey (1995, 1999) and continuously distributed in the rainshadow of recommended that only Desert Bighorn Sheep and the eastern slopes of the Continental Divide the Sierra Nevada population of California Bighorn throughout their range. Sheep be recognized as separate subspecies. California Bighorn Sheep were extirpated from most Currently, California and Rocky Mountain Bighorn of the United States by epizootic disease contracted sheep are managed as separate ecotypes in British from domestic sheep in the 1800s with a small Columbia. number living in California until 1954 (Buechner Description 1960). Since 1954, Bighorn Sheep have been reintroduced from British Columbia to California, California Bighorn Sheep are slightly smaller than Idaho, Nevada, North Dakota, Oregon, Utah, and mature Rocky Mountain Bighorn Sheep Washington, resulting in their re-establishment in (McTaggart-Cowan and Guiguet 1965). Like their much of their historic range. By 1998, California Rocky Mountain counterpart, California Bighorn Bighorn Sheep were estimated to number 10 000 Sheep have a dark to medium rich brown head, neck, (Toweill 1999). -

Bison Literature Review Biology
Bison Literature Review Ben Baldwin and Kody Menghini The purpose of this document is to compare the biology, ecology and basic behavior of cattle and bison for a management context. The literature related to bison is extensive and broad in scope covering the full continuum of domestication. The information incorporated in this review is focused on bison in more or less “wild” or free-ranging situations i.e.., not bison in close confinement or commercial production. While the scientific literature provides a solid basis for much of the basic biology and ecology, there is a wealth of information related to management implications and guidelines that is not captured. Much of the current information related to bison management, behavior (especially social organization) and practical knowledge is available through local experts, current research that has yet to be published, or popular literature. These sources, while harder to find and usually more localized in scope, provide crucial information pertaining to bison management. Biology Diet Composition Bison evolutional history provides the basis for many of the differences between bison and cattle. Bison due to their evolution in North America ecosystems are better adapted than introduced cattle, especially in grass dominated systems such as prairies. Many of these areas historically had relatively low quality forage. Bison are capable of more efficient digestion of low-quality forage then cattle (Peden et al. 1973; Plumb and Dodd 1993). Peden et al. (1973) also found that bison could consume greater quantities of low protein and poor quality forage then cattle. Bison and cattle have significant dietary overlap, but there are slight differences as well. -

Last Interglacial (MIS 5) Ungulate Assemblage from the Central Iberian Peninsula: the Camino Cave (Pinilla Del Valle, Madrid, Spain)
Palaeogeography, Palaeoclimatology, Palaeoecology 374 (2013) 327–337 Contents lists available at SciVerse ScienceDirect Palaeogeography, Palaeoclimatology, Palaeoecology journal homepage: www.elsevier.com/locate/palaeo Last Interglacial (MIS 5) ungulate assemblage from the Central Iberian Peninsula: The Camino Cave (Pinilla del Valle, Madrid, Spain) Diego J. Álvarez-Lao a,⁎, Juan L. Arsuaga b,c, Enrique Baquedano d, Alfredo Pérez-González e a Área de Paleontología, Departamento de Geología, Universidad de Oviedo, C/Jesús Arias de Velasco, s/n, 33005 Oviedo, Spain b Centro Mixto UCM-ISCIII de Evolución y Comportamiento Humanos, C/Sinesio Delgado, 4, 28029 Madrid, Spain c Departamento de Paleontología, Facultad de Ciencias Geológicas, Universidad Complutense de Madrid, Ciudad Universitaria, 28040 Madrid, Spain d Museo Arqueológico Regional de la Comunidad de Madrid, Plaza de las Bernardas, s/n, 28801-Alcalá de Henares, Madrid, Spain e Centro Nacional de Investigación sobre la Evolución Humana (CENIEH), Paseo Sierra de Atapuerca, s/n, 09002 Burgos, Spain article info abstract Article history: The fossil assemblage from the Camino Cave, corresponding to the late MIS 5, constitutes a key record to un- Received 2 November 2012 derstand the faunal composition of Central Iberia during the last Interglacial. Moreover, the largest Iberian Received in revised form 21 January 2013 fallow deer fossil population was recovered here. Other ungulate species present at this assemblage include Accepted 31 January 2013 red deer, roe deer, aurochs, chamois, wild boar, horse and steppe rhinoceros; carnivores and Neanderthals Available online 13 February 2013 are also present. The origin of the accumulation has been interpreted as a hyena den. Abundant fallow deer skeletal elements allowed to statistically compare the Camino Cave fossils with other Keywords: Early Late Pleistocene Pleistocene and Holocene European populations. -

Antelope, Deer, Bighorn Sheep and Mountain Goats: a Guide to the Carpals
J. Ethnobiol. 10(2):169-181 Winter 1990 ANTELOPE, DEER, BIGHORN SHEEP AND MOUNTAIN GOATS: A GUIDE TO THE CARPALS PAMELA J. FORD Mount San Antonio College 1100 North Grand Avenue Walnut, CA 91739 ABSTRACT.-Remains of antelope, deer, mountain goat, and bighorn sheep appear in archaeological sites in the North American west. Carpal bones of these animals are generally recovered in excellent condition but are rarely identified beyond the classification 1/small-sized artiodactyl." This guide, based on the analysis of over thirty modem specimens, is intended as an aid in the identifi cation of these remains for archaeological and biogeographical studies. RESUMEN.-Se han encontrado restos de antilopes, ciervos, cabras de las montanas rocosas, y de carneros cimarrones en sitios arqueol6gicos del oeste de Norte America. Huesos carpianos de estos animales se recuperan, por 10 general, en excelentes condiciones pero raramente son identificados mas alIa de la clasifi cacion "artiodactilos pequeno." Esta glia, basada en un anaIisis de mas de treinta especlmenes modemos, tiene el proposito de servir como ayuda en la identifica cion de estos restos para estudios arqueologicos y biogeogrMicos. RESUME.-On peut trouver des ossements d'antilopes, de cerfs, de chevres de montagne et de mouflons des Rocheuses, dans des sites archeologiques de la . region ouest de I'Amerique du Nord. Les os carpeins de ces animaux, generale ment en excellente condition, sont rarement identifies au dela du classement d' ,I artiodactyles de petite taille." Le but de ce guide base sur 30 specimens recents est d'aider aidentifier ces ossements pour des etudes archeologiques et biogeo graphiques. -

Genital Brucella Suis Biovar 2 Infection of Wild Boar (Sus Scrofa) Hunted in Tuscany (Italy)
microorganisms Article Genital Brucella suis Biovar 2 Infection of Wild Boar (Sus scrofa) Hunted in Tuscany (Italy) Giovanni Cilia * , Filippo Fratini , Barbara Turchi, Marta Angelini, Domenico Cerri and Fabrizio Bertelloni Department of Veterinary Science, University of Pisa, Viale delle Piagge 2, 56124 Pisa, Italy; fi[email protected] (F.F.); [email protected] (B.T.); [email protected] (M.A.); [email protected] (D.C.); [email protected] (F.B.) * Correspondence: [email protected] Abstract: Brucellosis is a zoonosis caused by different Brucella species. Wild boar (Sus scrofa) could be infected by some species and represents an important reservoir, especially for B. suis biovar 2. This study aimed to investigate the prevalence of Brucella spp. by serological and molecular assays in wild boar hunted in Tuscany (Italy) during two hunting seasons. From 287 animals, sera, lymph nodes, livers, spleens, and reproductive system organs were collected. Within sera, 16 (5.74%) were positive to both rose bengal test (RBT) and complement fixation test (CFT), with titres ranging from 1:4 to 1:16 (corresponding to 20 and 80 ICFTU/mL, respectively). Brucella spp. DNA was detected in four lymph nodes (1.40%), five epididymides (1.74%), and one fetus pool (2.22%). All positive PCR samples belonged to Brucella suis biovar 2. The results of this investigation confirmed that wild boar represents a host for B. suis biovar. 2 and plays an important role in the epidemiology of brucellosis in central Italy. Additionally, epididymis localization confirms the possible venereal transmission. Citation: Cilia, G.; Fratini, F.; Turchi, B.; Angelini, M.; Cerri, D.; Bertelloni, Keywords: Brucella suis biovar 2; wild boar; surveillance; epidemiology; reproductive system F. -

Anaplasma Phagocytophilum and Babesia Species Of
pathogens Article Anaplasma phagocytophilum and Babesia Species of Sympatric Roe Deer (Capreolus capreolus), Fallow Deer (Dama dama), Sika Deer (Cervus nippon) and Red Deer (Cervus elaphus) in Germany Cornelia Silaghi 1,2,*, Julia Fröhlich 1, Hubert Reindl 3, Dietmar Hamel 4 and Steffen Rehbein 4 1 Institute of Comparative Tropical Medicine and Parasitology, Ludwig-Maximilians-Universität München, Leopoldstr. 5, 80802 Munich, Germany; [email protected] 2 Institute of Infectology, Friedrich-Loeffler-Institut, Südufer 10, 17493 Greifswald Insel Riems, Germany 3 Tierärztliche Fachpraxis für Kleintiere, Schießtrath 12, 92709 Moosbach, Germany; [email protected] 4 Boehringer Ingelheim Vetmedica GmbH, Kathrinenhof Research Center, Walchenseestr. 8-12, 83101 Rohrdorf, Germany; [email protected] (D.H.); steff[email protected] (S.R.) * Correspondence: cornelia.silaghi@fli.de; Tel.: +49-0-383-5171-172 Received: 15 October 2020; Accepted: 18 November 2020; Published: 20 November 2020 Abstract: (1) Background: Wild cervids play an important role in transmission cycles of tick-borne pathogens; however, investigations of tick-borne pathogens in sika deer in Germany are lacking. (2) Methods: Spleen tissue of 74 sympatric wild cervids (30 roe deer, 7 fallow deer, 22 sika deer, 15 red deer) and of 27 red deer from a farm from southeastern Germany were analyzed by molecular methods for the presence of Anaplasma phagocytophilum and Babesia species. (3) Results: Anaplasma phagocytophilum and Babesia DNA was demonstrated in 90.5% and 47.3% of the 74 combined wild cervids and 14.8% and 18.5% of the farmed deer, respectively. Twelve 16S rRNA variants of A. phagocytophilum were delineated. -

SPR-659: Genetic Variation of Pronghorn Across US Route 89 And
Genetic Variation of Pronghorn across US Route 89 and State Route 64 Final Report 659 March 2012 Arizona Department of Transportation Research Center Genetic Variation of Pronghorn across US Route 89 and State Route 64 Final Report 659 March 2012 Prepared by: Tad Theimer, Scott Sprague, Ellyce Eddy, and Russell Benford Department of Biological Sciences Northern Arizona University Box 5640 Flagstaff, AZ 86011 Prepared for: Arizona Department of Transportation In cooperation with U.S. Department of Transportation Federal Highway Administration The contents of this report reflect the views of the authors who are responsible for the facts and the accuracy of the data presented herein. The contents do not necessarily reflect the official views or policies of the Arizona Department of Transportation or the Federal Highway Administration. This report does not constitute a standard, specification, or regulation. Trade or manufacturers’ names that may appear herein are cited only because they are considered essential to the objectives of the report. The US government and the State of Arizona do not endorse products or manufacturers. Cover photos courtesy of Wikipedia Commons. Technical Report Documentation Page 1. Report No. 2. Government Accession No. 3. Recipient's Catalog No. FHWA-AZ-12-659 4. Title and Subtitle 5. Report Date GENETIC VARIATION OF PRONGHORN ACROSS US ROUTE 89 AND March 2012 STATE ROUTE 64 6. Performing Organization Code 7. Author 8. Performing Organization Report No. Tad Theimer, Scott Sprague, Ellyce Eddy, Russell Benford 9. Performing Organization Name and Address 10. Work Unit No. Northern Arizona University Box 5640, Beaver Street Flagstaff, AZ 86011 11. -
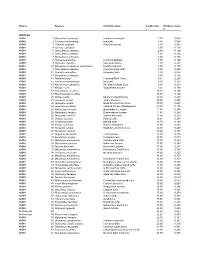
PDF File Containing Table of Lengths and Thicknesses of Turtle Shells And
Source Species Common name length (cm) thickness (cm) L t TURTLES AMNH 1 Sternotherus odoratus common musk turtle 2.30 0.089 AMNH 2 Clemmys muhlenbergi bug turtle 3.80 0.069 AMNH 3 Chersina angulata Angulate tortoise 3.90 0.050 AMNH 4 Testudo carbonera 6.97 0.130 AMNH 5 Sternotherus oderatus 6.99 0.160 AMNH 6 Sternotherus oderatus 7.00 0.165 AMNH 7 Sternotherus oderatus 7.00 0.165 AMNH 8 Homopus areolatus Common padloper 7.95 0.100 AMNH 9 Homopus signatus Speckled tortoise 7.98 0.231 AMNH 10 Kinosternon subrabum steinochneri Florida mud turtle 8.90 0.178 AMNH 11 Sternotherus oderatus Common musk turtle 8.98 0.290 AMNH 12 Chelydra serpentina Snapping turtle 8.98 0.076 AMNH 13 Sternotherus oderatus 9.00 0.168 AMNH 14 Hardella thurgi Crowned River Turtle 9.04 0.263 AMNH 15 Clemmys muhlenbergii Bog turtle 9.09 0.231 AMNH 16 Kinosternon subrubrum The Eastern Mud Turtle 9.10 0.253 AMNH 17 Kinixys crosa hinged-back tortoise 9.34 0.160 AMNH 18 Peamobates oculifers 10.17 0.140 AMNH 19 Peammobates oculifera 10.27 0.140 AMNH 20 Kinixys spekii Speke's hinged tortoise 10.30 0.201 AMNH 21 Terrapene ornata ornate box turtle 10.30 0.406 AMNH 22 Terrapene ornata North American box turtle 10.76 0.257 AMNH 23 Geochelone radiata radiated tortoise (Madagascar) 10.80 0.155 AMNH 24 Malaclemys terrapin diamondback terrapin 11.40 0.295 AMNH 25 Malaclemys terrapin Diamondback terrapin 11.58 0.264 AMNH 26 Terrapene carolina eastern box turtle 11.80 0.259 AMNH 27 Chrysemys picta Painted turtle 12.21 0.267 AMNH 28 Chrysemys picta painted turtle 12.70 0.168 AMNH 29 -

Evaluating How Swedish Hunters Determine Which Species Belong in Nature
European Journal of Wildlife Research (2020) 66: 77 https://doi.org/10.1007/s10344-020-01418-6 ORIGINAL ARTICLE Evaluating how Swedish hunters determine which species belong in nature M. Nils Peterson1 & Alyssa Chen1 & Erica von Essen1 & Hans Peter Hansen1 Received: 30 January 2020 /Revised: 17 August 2020 /Accepted: 24 August 2020 / Published online: 27 August 2020 # Springer-Verlag GmbH Germany, part of Springer Nature 2020 Abstract Understanding whether people view non-native species as belonging in a place will help guide important conservation efforts ranging from eradications of exotics to re-introduction of extirpated species. In this manuscript we describe the degree to which Swedish hunters perceive key wildlife species as belonging in Swedish nature. We surveyed 2014 Swedish hunters randomly selected from a database of all registered hunters with a 47.5% response rate. We measured hunters’ perceptions of the belonging of 10 key species on the Swedish landscape, compared them with confidence intervals for proportions, and predicted them using regression models. Construct validity was assessed through pretesting and focus groups. Our results suggest Swedish hunters consider species introduced wholly by humans as less likely to belong in Sweden compared with species that evolved in situ, species with negative socio-economic impact as less likely to belong in Sweden compared with species with no impact or positive economic impacts, and species with wide distributions to be seen as more likely to belong in Sweden compared with those with narrow distributions. Perceptions of wolves, fallow deer, and European rabbits differed from these broad trends potentially due to unique cultural constructions of belonging for the species and the duration since anthropogenic introductions for the latter species. -

Preliminary Studies on the Etiology of Keratoconjunctivitis in Reindeer (Rangifer Tarandus Tarandus) Calves in Alaska
Journal of Wildlife Diseases, 44(4), 2008, pp. 1051–1055 # Wildlife Disease Association 2008 Preliminary Studies on the Etiology of Keratoconjunctivitis in Reindeer (Rangifer tarandus tarandus) Calves in Alaska Alina L. Evans,1,5 Russell F. Bey,1 James V. Schoster,2 James E. Gaarder,3 and Gregory L. Finstad4 1 Department of Veterinary and Biomedical Sciences, College of Veterinary Medicine, University of Minnesota, 1971 Commonwealth Ave., St. Paul, Minnesota 55108, USA; 2 Animal Eye Consultants of Minnesota, Roseville, Minnesota 55113, USA; 3 Veterinary Eye Specialists, 1921 W Diamond Blvd., Suite 108, Anchorage, Alaska, 99515, USA; 4 Reindeer Research Program, University of Alaska, PO Box 757200, Fairbanks, Alaska, 99775; 5 Corresponding author (email: [email protected]) ABSTRACT: Keratoconjunctivitis outbreaks oc- and possibly contagious eye disease that cur each summer in reindeer (Rangifer tar- can leave animals blind or with impaired andus tarandus) herds in western Alaska, USA. vision. Keratoconjunctivitis is seen annu- This condition has not been well characterized nor has a definitive primary etiologic agent ally during the summer reindeer handlings been identified. We evaluated the eyes of 660 on the Seward Peninsula (Reindeer Re- calves near Nome, Alaska, between 29 June and search Program, University of Alaska 14 July 2005. Clinical signs of keratoconjuncti- Fairbanks, unpubl. data). vitis were observed in 26/660 calves (3.9%). Infectious keratoconjunctivitis has been Samples were collected from the conjunctival studied in numerous other species. In sac of both affected (n522) and unaffected (n524) animals for bacterial culture, enzyme- cattle, the primary pathogen has been linked immunosorbent assay testing for Chla- identified to be the piliated form of mydophila psittaci, and for polymerase chain Moraxella bovis (Ruehl et al., 1988). -

European Bison
IUCN/Species Survival Commission Status Survey and Conservation Action Plan The Species Survival Commission (SSC) is one of six volunteer commissions of IUCN – The World Conservation Union, a union of sovereign states, government agencies and non- governmental organisations. IUCN has three basic conservation objectives: to secure the conservation of nature, and especially of biological diversity, as an essential foundation for the future; to ensure that where the Earth’s natural resources are used this is done in a wise, European Bison equitable and sustainable way; and to guide the development of human communities towards ways of life that are both of good quality and in enduring harmony with other components of the biosphere. A volunteer network comprised of some 8,000 scientists, field researchers, government officials Edited by Zdzis³aw Pucek and conservation leaders from nearly every country of the world, the SSC membership is an Compiled by Zdzis³aw Pucek, Irina P. Belousova, unmatched source of information about biological diversity and its conservation. As such, SSC Ma³gorzata Krasiñska, Zbigniew A. Krasiñski and Wanda Olech members provide technical and scientific counsel for conservation projects throughout the world and serve as resources to governments, international conventions and conservation organisations. IUCN/SSC Action Plans assess the conservation status of species and their habitats, and specifies conservation priorities. The series is one of the world’s most authoritative sources of species conservation information -

Phylogeny of Dictyocaulus (Lungworms) from Eight Species of Ruminants Based on Analyses of Ribosomal RNA Data
179 Phylogeny of Dictyocaulus (lungworms) from eight species of ruminants based on analyses of ribosomal RNA data J. HO¨ GLUND1*,D.A.MORRISON1, B. P. DIVINA1,2, E. WILHELMSSON1 and J. G. MATTSSON1 1 Department of Parasitology (SWEPAR), National Veterinary Institute and Swedish University of Agriculture Sciences, 751 89 Uppsala, Sweden 2 College of Veterinary Medicine, University of the Philippines Los Ban˜os, College, Laguna, 4031 Philippines (Received 8 November 2002; revised 8 February 2003; accepted 8 February 2003) SUMMARY In this study, we conducted phylogenetic analyses of nematode parasites within the genus Dictyocaulus (superfamily Trichostrongyloidea). Lungworms from cattle (Bos taurus), domestic sheep (Ovis aries), European fallow deer (Dama dama), moose (Alces alces), musk ox (Ovibos moschatus), red deer (Cervus elaphus), reindeer (Rangifer tarandus) and roe deer (Capreolus capreolus) were obtained and their small subunit ribosomal RNA (SSU) and internal transcribed spacer 2 (ITS2) sequences analysed. In the hosts examined we identified D. capreolus, D. eckerti, D. filaria and D. viviparus. However, in fallow deer we detected a taxon with unique SSU and ITS2 sequences. The phylogenetic position of this taxon based on the SSU sequences shows that it is a separate evolutionary lineage from the other recognized species of Dictyocaulus. Furthermore, the analysis of the ITS2 sequence data indicates that it is as genetically distinct as are the named species of Dictyocaulus. Therefore, either this taxon needs to be recognized as a new species, or D. capreolus, D. eckerti and D. viviparus need to be combined into a single species. Traditionally, the genus Dictyocaulus has been placed as a separate family within the superfamily Trichostrongyloidea.