INFORMATION to USERS This Manuscript Has Been Reproduced from the Microfilm Master
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Arabian Ungulate CAMP & Leopard, Tahr, and Oryx PHVA Final Report 2001.Pdf
Conservation Assessment and Management Plan (CAMP) For The Arabian Ungulates and Leopard & Population and Habitat Viability Assessment (PHVA) For the Arabian Leopard, Tahr, and Arabian Oryx 1 © Copyright 2001 by CBSG. A contribution of the IUCN/SSC Conservation Breeding Specialist Group. Conservation Breeding Specialist Group (SSC/IUCN). 2001. Conservation Assessment and Management Plan for the Arabian Leopard and Arabian Ungulates with Population and Habitat Viability Assessments for the Arabian Leopard, Arabian Oryx, and Tahr Reports. CBSG, Apple Valley, MN. USA. Additional copies of Conservation Assessment and Management Plan for the Arabian Leopard and Arabian Ungulates with Population and Habitat Viability Assessments for the Arabian Leopard, Arabian Oryx, and Tahr Reports can be ordered through the IUCN/SSC Conservation Breeding Specialist Group, 12101 Johnny Cake Ridge Road, Apple Valley, MN 55124. USA. 2 Donor 3 4 Conservation Assessment and Management Plan (CAMP) For The Arabian Ungulates and Leopard & Population and Habitat Viability Assessment (PHVA) For the Arabian Leopard, Tahr, and Arabian Oryx TABLE OF CONTENTS SECTION 1: Executive Summary 5. SECTION 2: Arabian Gazelles Reports 18. SECTION 3: Tahr and Ibex Reports 28. SECTION 4: Arabian Oryx Reports 41. SECTION 5: Arabian Leopard Reports 56. SECTION 6: New IUCN Red List Categories & Criteria; Taxon Data Sheet; and CBSG Workshop Process. 66. SECTION 7: List of Participants 116. 5 6 Conservation Assessment and Management Plan (CAMP) For The Arabian Ungulates and Leopard & Population and Habitat Viability Assessment (PHVA) For the Arabian Leopard, Tahr, and Arabian Oryx SECTION 1 Executive Summary 7 8 Executive Summary The ungulates of the Arabian peninsula region - Arabian Oryx, Arabian tahr, ibex, and the gazelles - generally are poorly known among local communities and the general public. -

08E – CHAMOIS (GAMSWILD)
U.S. Forces Europe Hunting, Fishing, and Sport Shooting Program Guide to Hunting in Germany 08e – CHAMOIS (GAMSWILD) Breeding season (Brunftzeit): November Gestation period: 20-21 weeks Young are born: May (one, sometimes two) Teeth: 32 Males: Gamsbock Females: Geiss Fawns: Kitz Herd: Rudel Based on fossils, scientists believe that some 100,000 years ago, ancestors of chamois known in Europe today originally migrated from Central Asia. These populations were wiped out during the last ice-age, except for chamois that are located in the Apennine and the Pyrenees mountain ranges. When the glaciers melted away, about 10,000 years ago, another Chamois migration from Asia led to a re-population of Eastern and Central Europe, except for Scandinavia and the British Islands. 08e-1 U.S. Forces Europe Hunting, Fishing, and Sport Shooting Program Guide to Hunting in Germany As a true mountain animal, Chamois lived mainly in regions with rocks and steep cliffs and outside the forest. With the increasing extension of forest on the former taiga landscape, Chamois became restricted to Alpine areas. In this sense, “Alpine” means regions above the forest line. These were mainly the Carpathians, the Tatras, the Dinarids, the Alps, the Apennines, and the Pyrenees. Aside from alpine regions, Chamois have also been introduced successfully to some parts in Europe (Schwarzwald and Elbsandsteingebirge, Germany), and to New Zealand. The chamois is a member of the family Bovidae. Chamois is a very distinct genus, with no close relatives in the world of cloven hoofed game. The Serau and the Goral, both in Central Asia, and the Snow Goat in North America are considered the closest relatives. -

Last Interglacial (MIS 5) Ungulate Assemblage from the Central Iberian Peninsula: the Camino Cave (Pinilla Del Valle, Madrid, Spain)
Palaeogeography, Palaeoclimatology, Palaeoecology 374 (2013) 327–337 Contents lists available at SciVerse ScienceDirect Palaeogeography, Palaeoclimatology, Palaeoecology journal homepage: www.elsevier.com/locate/palaeo Last Interglacial (MIS 5) ungulate assemblage from the Central Iberian Peninsula: The Camino Cave (Pinilla del Valle, Madrid, Spain) Diego J. Álvarez-Lao a,⁎, Juan L. Arsuaga b,c, Enrique Baquedano d, Alfredo Pérez-González e a Área de Paleontología, Departamento de Geología, Universidad de Oviedo, C/Jesús Arias de Velasco, s/n, 33005 Oviedo, Spain b Centro Mixto UCM-ISCIII de Evolución y Comportamiento Humanos, C/Sinesio Delgado, 4, 28029 Madrid, Spain c Departamento de Paleontología, Facultad de Ciencias Geológicas, Universidad Complutense de Madrid, Ciudad Universitaria, 28040 Madrid, Spain d Museo Arqueológico Regional de la Comunidad de Madrid, Plaza de las Bernardas, s/n, 28801-Alcalá de Henares, Madrid, Spain e Centro Nacional de Investigación sobre la Evolución Humana (CENIEH), Paseo Sierra de Atapuerca, s/n, 09002 Burgos, Spain article info abstract Article history: The fossil assemblage from the Camino Cave, corresponding to the late MIS 5, constitutes a key record to un- Received 2 November 2012 derstand the faunal composition of Central Iberia during the last Interglacial. Moreover, the largest Iberian Received in revised form 21 January 2013 fallow deer fossil population was recovered here. Other ungulate species present at this assemblage include Accepted 31 January 2013 red deer, roe deer, aurochs, chamois, wild boar, horse and steppe rhinoceros; carnivores and Neanderthals Available online 13 February 2013 are also present. The origin of the accumulation has been interpreted as a hyena den. Abundant fallow deer skeletal elements allowed to statistically compare the Camino Cave fossils with other Keywords: Early Late Pleistocene Pleistocene and Holocene European populations. -

Status and Protection of Globally Threatened Species in the Caucasus
STATUS AND PROTECTION OF GLOBALLY THREATENED SPECIES IN THE CAUCASUS CEPF Biodiversity Investments in the Caucasus Hotspot 2004-2009 Edited by Nugzar Zazanashvili and David Mallon Tbilisi 2009 The contents of this book do not necessarily reflect the views or policies of CEPF, WWF, or their sponsoring organizations. Neither the CEPF, WWF nor any other entities thereof, assumes any legal liability or responsibility for the accuracy, completeness, or usefulness of any information, product or process disclosed in this book. Citation: Zazanashvili, N. and Mallon, D. (Editors) 2009. Status and Protection of Globally Threatened Species in the Caucasus. Tbilisi: CEPF, WWF. Contour Ltd., 232 pp. ISBN 978-9941-0-2203-6 Design and printing Contour Ltd. 8, Kargareteli st., 0164 Tbilisi, Georgia December 2009 The Critical Ecosystem Partnership Fund (CEPF) is a joint initiative of l’Agence Française de Développement, Conservation International, the Global Environment Facility, the Government of Japan, the MacArthur Foundation and the World Bank. This book shows the effort of the Caucasus NGOs, experts, scientific institutions and governmental agencies for conserving globally threatened species in the Caucasus: CEPF investments in the region made it possible for the first time to carry out simultaneous assessments of species’ populations at national and regional scales, setting up strategies and developing action plans for their survival, as well as implementation of some urgent conservation measures. Contents Foreword 7 Acknowledgments 8 Introduction CEPF Investment in the Caucasus Hotspot A. W. Tordoff, N. Zazanashvili, M. Bitsadze, K. Manvelyan, E. Askerov, V. Krever, S. Kalem, B. Avcioglu, S. Galstyan and R. Mnatsekanov 9 The Caucasus Hotspot N. -

Capture, Restraint and Transport Stress in Southern Chamois (Rupicapra Pyrenaica)
Capture,Capture, restraintrestraint andand transporttransport stressstress ininin SouthernSouthern chamoischamois ((RupicapraRupicapra pyrenaicapyrenaica)) ModulationModulation withwith acepromazineacepromazine andand evaluationevaluation usingusingusing physiologicalphysiologicalphysiological parametersparametersparameters JorgeJorgeJorge RamónRamónRamón LópezLópezLópez OlveraOlveraOlvera 200420042004 Capture, restraint and transport stress in Southern chamois (Rupicapra pyrenaica) Modulation with acepromazine and evaluation using physiological parameters Jorge Ramón López Olvera Bellaterra 2004 Esta tesis doctoral fue realizada gracias a la financiación de la Comisión Interministerial de Ciencia y Tecnología (proyecto CICYT AGF99- 0763-C02) y a una beca predoctoral de Formación de Investigadores de la Universidad Autónoma de Barcelona, y contó con el apoyo del Departament de Medi Ambient de la Generalitat de Catalunya. Los Doctores SANTIAGO LAVÍN GONZÁLEZ e IGNASI MARCO SÁNCHEZ, Catedrático de Universidad y Profesor Titular del Área de Conocimiento de Medicina y Cirugía Animal de la Facultad de Veterinaria de la Universidad Autónoma de Barcelona, respectivamente, CERTIFICAN: Que la memoria titulada ‘Capture, restraint and transport stress in Southern chamois (Rupicapra pyrenaica). Modulation with acepromazine and evaluation using physiological parameters’, presentada por el licenciado Don JORGE R. LÓPEZ OLVERA para la obtención del grado de Doctor en Veterinaria, se ha realizado bajo nuestra dirección y, considerándola satisfactoriamente -

SPANISH+IBEX+PACKAGE.Pdf
La Pedriza Caza y Gestion is operated by his Spanish Professional Hunter & Outfitter Pedro Alarcón, since 1999, Pedro born into a family with very strong hunting tradition, in addition, this passion for hunting pushed to study forest engineering, expanding his knowledge of the game and his enviroments. Our home is in Cordoba, the south of Spain. We are specializes in big game hunt throughout the all our landscape, mainly in (free range-no fence area) private concessions & goberment areas. For Spanish Ibex we have 4 species (Beceite, Gredos, Southeastern & Ronda), Spanish Red Stag, European Fallow Deer, Mouflon Sheep, Pyrenean Chamois, Cantabrian Chamois, European Roe Deer, Barbary Sheep or Aoudad and Wild Boar. In our areas many time we get record trophies. Also we are specialist in Monterias and driven partridge. HUNTING AREA: Beceite, Gredos, Ronda and South East. (Free Range, no fence area) WAY OF HUNTING: Stalking TIME: Any time GROUP SIZE: 2-3 hunters ACCOMMODATION: Luxury hotels in hunting area. I T I N E R A R Y Day 1 Arrival at the airport (Madrid, Valencia or Granada) Day 2-4 Hunting (depend animals) Day 5 Back at the airport and departure. If you want the ibex grand sland or more different animals, we need about 3 days per animals, plus 1 day between hunting areas. SPECIAL PACKAGE PRICE 3.950€ 1 X 1 5 Days trip with a Beceite or Ronda or South Eastern Ibex included 3 Hunting days and 2 travel days in luxury accommodation and all meal included. The trophy fee of the ibex up to 205CIC points (bronce medals class). -

Arabian Tahr in Oman Paul Munton
Arabian Tahr in Oman Paul Munton Arabian tahr are confined to Oman, with a population of under 2000. Unlike other tahr species, which depend on grass, Arabian tahr require also fruits, seeds and young shoots. The areas where these can be found in this arid country are on certain north-facing mountain slopes with a higher rainfall, and it is there that reserves to protect this tahr must be made. The author spent two years in Oman studying the tahr. The Arabian tahr Hemitragus jayakari today survives only in the mountains of northern Oman. A goat-like animal, it is one of only three surviving species of a once widespread genus; the other two are the Himalayan and Nilgiri tahrs, H. jemlahicus and H. hylocrius. In recent years the government of the Sultanate of Oman has shown great interest in the country's wildlife, and much conservation work has been done. From April 1976 to April 1978 I was engaged jointly by the Government, WWF and IUCN on a field study of the tahr's ecology, and in January 1979 made recommendations for its conservation, which were presented to the Government. Arabian tahr differ from the other tahrs in that they feed selectively on fruits, seeds and young shoots as well as grass. Their optimum habitat is found on the north-facing slopes of the higher mountain ranges of northern Oman, where they use all altitudes between sea level and 2000 metres. But they prefer the zone between 1000 and 1800m where the vegetation is especially diverse, due to the special climate of these north-facing slopes, with their higher rainfall, cooler temperatures, and greater shelter from the sun than in the drought conditions that are otherwise typical of this arid zone. -

Central Eurasian Aridland Mammals Action Plan
CMS CONVENTION ON Distr. General MIGRATORY UNEP/CMS/ScC17/Doc.13 SPECIES 8 November 2011 Original: English 17 th MEETING OF THE SCIENTIFIC COUNCIL Bergen, 17-18 November 2011 Agenda Item 17.3.6 CENTRAL EURASIAN ARIDLAND MAMMALS ACTION PLAN (Prepared by the Secretariat) Following COP Recommendation 9.1 the Secretariat has prepared a draft Action Plan to complement the Concerted and Cooperative Action for Central Eurasian Aridland Mammals. The document is a first draft, intended to stimulate discussion and identify further action needed to finalize the document in consultation with the Range States and other stakeholders, and to agree on next steps towards its implementation. Action requested: The 17 th Meeting of the Scientific Council is invited to: a. Take note of the document and provide guidance on its further development and implementation; b. Review and advise in particular on the definition of the geographic scope, including the range states, and the target species (listed in table 1); and c. Provide guidance on the terminology currently used for the Action Plan, agree on a definition of the term aridlands and/or consider using the term drylands instead. Central Eurasian Aridland Mammals Draft Action Plan Produced by the UNEP/CMS Secretariat November 2011 1 Content 1. Introduction ................................................................................................................... 3 1.1 Vision and Main Priority Directions ................................................................................................... -

Towards Snow Leopard Prey Recovery: Understanding the Resource Use Strategies and Demographic Responses of Bharal Pseudois Nayaur to Livestock Grazing and Removal
Towards snow leopard prey recovery: understanding the resource use strategies and demographic responses of bharal Pseudois nayaur to livestock grazing and removal Final project report submitted by Kulbhushansingh Suryawanshi Nature Conservation Foundation, Mysore Post-graduate Program in Wildlife Biology and Conservation, National Centre for Biological Sciences, Wildlife Conservation Society –India program, Bangalore, India To Snow Leopard Conservation Grant Program January 2009 Towards snow leopard prey recovery: understanding the resource use strategies and demographic responses of bharal Pseudois nayaur to livestock grazing and removal. 1. Executive Summary: Decline of wild prey populations in the Himalayan region, largely due to competition with livestock, has been identified as one of the main threats to the snow leopard Uncia uncia. Studies show that bharal Pseudois nayaur diet is dominated by graminoids during summer, but the proportion of graminoids declines in winter. We explore the causes for the decline of graminoids from bharal winter diet and resulting implications for bharal conservation. We test the predictions generated by two alternative hypotheses, (H1) low graminoid availability caused by livestock grazing during winter causes bharal to include browse in their diet, and, (H2) bharal include browse, with relatively higher nutrition, to compensate for the poor quality of graminoids during winter. Graminoid availability was highest in areas without livestock grazing, followed by areas with moderate and intense livestock grazing. Graminoid quality in winter was relatively lower than that of browse, but the difference was not statistically significant. Bharal diet was dominated by graminoids in areas with highest graminoid availability. Graminoid contribution to bharal diet declined monotonically with a decline in graminoid availability. -

Field Guide Mammals of Ladakh ¾-Hðgå-ÅÛ-Hýh-ºiô-;Ým-Mû-Ç+Ô¼-¾-Zçàz-Çeômü
Field Guide Mammals of Ladakh ¾-hÐGÅ-ÅÛ-hÝh-ºIô-;Ým-mÛ-Ç+ô¼-¾-zÇÀz-Çeômü Tahir Shawl Jigmet Takpa Phuntsog Tashi Yamini Panchaksharam 2 FOREWORD Ladakh is one of the most wonderful places on earth with unique biodiversity. I have the privilege of forwarding the fi eld guide on mammals of Ladakh which is part of a series of bilingual (English and Ladakhi) fi eld guides developed by WWF-India. It is not just because of my involvement in the conservation issues of the state of Jammu & Kashmir, but I am impressed with the Ladakhi version of the Field Guide. As the Field Guide has been specially produced for the local youth, I hope that the Guide will help in conserving the unique mammal species of Ladakh. I also hope that the Guide will become a companion for every nature lover visiting Ladakh. I commend the efforts of the authors in bringing out this unique publication. A K Srivastava, IFS Chief Wildlife Warden, Govt. of Jammu & Kashmir 3 ÇSôm-zXôhü ¾-hÐGÅ-mÛ-ºWÛG-dïm-mP-¾-ÆôG-VGÅ-Ço-±ôGÅ-»ôh-źÛ-GmÅ-Å-h¤ÛGÅ-zž-ŸÛG-»Ûm-môGü ¾-hÐGÅ-ÅÛ-Å-GmÅ-;Ým-¾-»ôh-qºÛ-Åï¤Å-Tm-±P-¤ºÛ-MãÅ-‚Å-q-ºhÛ-¾-ÇSôm-zXôh-‚ô-‚Å- qôºÛ-PºÛ-¾Å-ºGm-»Ûm-môGü ºÛ-zô-P-¼P-W¤-¤Þ-;-ÁÛ-¤Û¼-¼Û-¼P-zŸÛm-D¤-ÆâP-Bôz-hP- ºƒï¾-»ôh-¤Dm-qôÅ-‚Å-¼ï-¤m-q-ºÛ-zô-¾-hÐGÅ-ÅÛ-Ç+h-hï-mP-P-»ôh-‚Å-qôº-È-¾Å-bï-»P- zÁh- »ôPÅü Åï¤Å-Tm-±P-¤ºÛ-MãÅ-‚ô-‚Å-qô-h¤ÛGÅ-zž-¾ÛÅ-GŸôm-mÝ-;Ým-¾-wm-‚Å-¾-ºwÛP-yï-»Ûm- môG ºô-zôºÛ-;-mÅ-¾-hÐGÅ-ÅÛ-h¤ÛGÅ-zž-Tm-mÛ-Åï¤Å-Tm-ÆâP-BôzÅ-¾-wm-qºÛ-¼Û-zô-»Ûm- hôm-m-®ôGÅ-¾ü ¼P-zŸÛm-D¤Å-¾-ºfh-qô-»ôh-¤Dm-±P-¤-¾ºP-wm-fôGÅ-qºÛ-¼ï-z-»Ûmü ºhÛ-®ßGÅ-ºô-zM¾-¤²h-hï-ºƒÛ-¤Dm-mÛ-ºhÛ-hqï-V-zô-q¼-¾-zMz-Çeï-Çtï¾-hGôÅ-»Ûm-môG Íï-;ï-ÁÙÛ-¶Å-b-z-ͺÛ-Íïw-ÍôÅ- mGÅ-±ôGÅ-Åï¤Å-Tm-ÆâP-Bôz-Çkï-DG-GÛ-hqôm-qô-G®ô-zô-W¤- ¤Þ-;ÁÛ-¤Û¼-GŸÝP.ü 4 5 ACKNOWLEDGEMENTS The fi eld guide is the result of exhaustive work by a large number of people. -
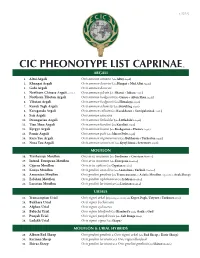
Cic Pheonotype List Caprinae©
v. 5.25.12 CIC PHEONOTYPE LIST CAPRINAE © ARGALI 1. Altai Argali Ovis ammon ammon (aka Altay Argali) 2. Khangai Argali Ovis ammon darwini (aka Hangai & Mid Altai Argali) 3. Gobi Argali Ovis ammon darwini 4. Northern Chinese Argali - extinct Ovis ammon jubata (aka Shansi & Jubata Argali) 5. Northern Tibetan Argali Ovis ammon hodgsonii (aka Gansu & Altun Shan Argali) 6. Tibetan Argali Ovis ammon hodgsonii (aka Himalaya Argali) 7. Kuruk Tagh Argali Ovis ammon adametzi (aka Kuruktag Argali) 8. Karaganda Argali Ovis ammon collium (aka Kazakhstan & Semipalatinsk Argali) 9. Sair Argali Ovis ammon sairensis 10. Dzungarian Argali Ovis ammon littledalei (aka Littledale’s Argali) 11. Tian Shan Argali Ovis ammon karelini (aka Karelini Argali) 12. Kyrgyz Argali Ovis ammon humei (aka Kashgarian & Hume’s Argali) 13. Pamir Argali Ovis ammon polii (aka Marco Polo Argali) 14. Kara Tau Argali Ovis ammon nigrimontana (aka Bukharan & Turkestan Argali) 15. Nura Tau Argali Ovis ammon severtzovi (aka Kyzyl Kum & Severtzov Argali) MOUFLON 16. Tyrrhenian Mouflon Ovis aries musimon (aka Sardinian & Corsican Mouflon) 17. Introd. European Mouflon Ovis aries musimon (aka European Mouflon) 18. Cyprus Mouflon Ovis aries ophion (aka Cyprian Mouflon) 19. Konya Mouflon Ovis gmelini anatolica (aka Anatolian & Turkish Mouflon) 20. Armenian Mouflon Ovis gmelini gmelinii (aka Transcaucasus or Asiatic Mouflon, regionally as Arak Sheep) 21. Esfahan Mouflon Ovis gmelini isphahanica (aka Isfahan Mouflon) 22. Larestan Mouflon Ovis gmelini laristanica (aka Laristan Mouflon) URIALS 23. Transcaspian Urial Ovis vignei arkal (Depending on locality aka Kopet Dagh, Ustyurt & Turkmen Urial) 24. Bukhara Urial Ovis vignei bocharensis 25. Afghan Urial Ovis vignei cycloceros 26. -

Reproductive Seasonality in Captive Wild Ruminants: Implications for Biogeographical Adaptation, Photoperiodic Control, and Life History
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2012 Reproductive seasonality in captive wild ruminants: implications for biogeographical adaptation, photoperiodic control, and life history Zerbe, Philipp Abstract: Zur quantitativen Beschreibung der Reproduktionsmuster wurden Daten von 110 Wildwiederkäuer- arten aus Zoos der gemässigten Zone verwendet (dabei wurde die Anzahl Tage, an denen 80% aller Geburten stattfanden, als Geburtenpeak-Breite [BPB] definiert). Diese Muster wurden mit verschiede- nen biologischen Charakteristika verknüpft und mit denen von freilebenden Tieren verglichen. Der Bre- itengrad des natürlichen Verbreitungsgebietes korreliert stark mit dem in Menschenobhut beobachteten BPB. Nur 11% der Spezies wechselten ihr reproduktives Muster zwischen Wildnis und Gefangenschaft, wobei für saisonale Spezies die errechnete Tageslichtlänge zum Zeitpunkt der Konzeption für freilebende und in Menschenobhut gehaltene Populationen gleich war. Reproduktive Saisonalität erklärt zusätzliche Varianzen im Verhältnis von Körpergewicht und Tragzeit, wobei saisonalere Spezies für ihr Körpergewicht eine kürzere Tragzeit aufweisen. Rückschliessend ist festzuhalten, dass Photoperiodik, speziell die abso- lute Tageslichtlänge, genetisch fixierter Auslöser für die Fortpflanzung ist, und dass die Plastizität der Tragzeit unterstützend auf die erfolgreiche Verbreitung der Wiederkäuer in höheren Breitengraden wirkte. A dataset on 110 wild ruminant species kept in captivity in temperate-zone zoos was used to describe their reproductive patterns quantitatively (determining the birth peak breadth BPB as the number of days in which 80% of all births occur); then this pattern was linked to various biological characteristics, and compared with free-ranging animals. Globally, latitude of natural origin highly correlates with BPB observed in captivity, with species being more seasonal originating from higher latitudes.