Untangling the Evolutionary History of European Bison (Bison Bonasus) Ayla L
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Bison Literature Review Biology
Bison Literature Review Ben Baldwin and Kody Menghini The purpose of this document is to compare the biology, ecology and basic behavior of cattle and bison for a management context. The literature related to bison is extensive and broad in scope covering the full continuum of domestication. The information incorporated in this review is focused on bison in more or less “wild” or free-ranging situations i.e.., not bison in close confinement or commercial production. While the scientific literature provides a solid basis for much of the basic biology and ecology, there is a wealth of information related to management implications and guidelines that is not captured. Much of the current information related to bison management, behavior (especially social organization) and practical knowledge is available through local experts, current research that has yet to be published, or popular literature. These sources, while harder to find and usually more localized in scope, provide crucial information pertaining to bison management. Biology Diet Composition Bison evolutional history provides the basis for many of the differences between bison and cattle. Bison due to their evolution in North America ecosystems are better adapted than introduced cattle, especially in grass dominated systems such as prairies. Many of these areas historically had relatively low quality forage. Bison are capable of more efficient digestion of low-quality forage then cattle (Peden et al. 1973; Plumb and Dodd 1993). Peden et al. (1973) also found that bison could consume greater quantities of low protein and poor quality forage then cattle. Bison and cattle have significant dietary overlap, but there are slight differences as well. -

Last Interglacial (MIS 5) Ungulate Assemblage from the Central Iberian Peninsula: the Camino Cave (Pinilla Del Valle, Madrid, Spain)
Palaeogeography, Palaeoclimatology, Palaeoecology 374 (2013) 327–337 Contents lists available at SciVerse ScienceDirect Palaeogeography, Palaeoclimatology, Palaeoecology journal homepage: www.elsevier.com/locate/palaeo Last Interglacial (MIS 5) ungulate assemblage from the Central Iberian Peninsula: The Camino Cave (Pinilla del Valle, Madrid, Spain) Diego J. Álvarez-Lao a,⁎, Juan L. Arsuaga b,c, Enrique Baquedano d, Alfredo Pérez-González e a Área de Paleontología, Departamento de Geología, Universidad de Oviedo, C/Jesús Arias de Velasco, s/n, 33005 Oviedo, Spain b Centro Mixto UCM-ISCIII de Evolución y Comportamiento Humanos, C/Sinesio Delgado, 4, 28029 Madrid, Spain c Departamento de Paleontología, Facultad de Ciencias Geológicas, Universidad Complutense de Madrid, Ciudad Universitaria, 28040 Madrid, Spain d Museo Arqueológico Regional de la Comunidad de Madrid, Plaza de las Bernardas, s/n, 28801-Alcalá de Henares, Madrid, Spain e Centro Nacional de Investigación sobre la Evolución Humana (CENIEH), Paseo Sierra de Atapuerca, s/n, 09002 Burgos, Spain article info abstract Article history: The fossil assemblage from the Camino Cave, corresponding to the late MIS 5, constitutes a key record to un- Received 2 November 2012 derstand the faunal composition of Central Iberia during the last Interglacial. Moreover, the largest Iberian Received in revised form 21 January 2013 fallow deer fossil population was recovered here. Other ungulate species present at this assemblage include Accepted 31 January 2013 red deer, roe deer, aurochs, chamois, wild boar, horse and steppe rhinoceros; carnivores and Neanderthals Available online 13 February 2013 are also present. The origin of the accumulation has been interpreted as a hyena den. Abundant fallow deer skeletal elements allowed to statistically compare the Camino Cave fossils with other Keywords: Early Late Pleistocene Pleistocene and Holocene European populations. -

Infectious Diseases of Saiga Antelopes and Domestic Livestock in Kazakhstan
Infectious diseases of saiga antelopes and domestic livestock in Kazakhstan Monica Lundervold University of Warwick, UK June 2001 1 Chapter 1 Introduction This thesis combines an investigation of the ecology of a wild ungulate, the saiga antelope (Saiga tatarica, Pallas), with epidemiological work on the diseases that this species shares with domestic livestock. The main focus is on foot-and-mouth disease (FMD) and brucellosis. The area of study was Kazakhstan (located in Central Asia, Figure 1.1), home to the largest population of saiga antelope in the world (Bekenov et al., 1998). Kazakhstan's independence from the Soviet Union in 1991 led to a dramatic economic decline, accompanied by a massive reduction in livestock numbers and a virtual collapse in veterinary services (Goskomstat, 1996; Morin, 1998a). As the rural economy has disintegrated, the saiga has suffered a dramatic increase in poaching (Bekenov et al., 1998). Thus the investigation reported in this thesis includes ecological, epidemiological and socio-economic aspects, all of which were necessary in order to gain a full picture of the dynamics of the infectious diseases of saigas and livestock in Kazakhstan. The saiga is an interesting species to study because it is one of the few wildlife populations in the world that has been successfully managed for commercial hunting over a period of more than 40 years (Milner-Gulland, 1994a). Its location in Central Asia, an area that was completely closed to foreigners during the Soviet era, means that very little information on the species and its management has been available in western literature. The diseases that saigas share with domestic livestock have been a particular focus of this study because of the interesting issues related to veterinary care and disease control in the Former Soviet Union (FSU). -

A Middle Holocene Steppe Bison and Paleoenvironments from the Versleuce Meadows, Whitehorse, Yukon, Canada Grant D
1138 ARTICLE A middle Holocene steppe bison and paleoenvironments from the Versleuce Meadows, Whitehorse, Yukon, Canada Grant D. Zazula, Elizabeth Hall, P. Gregory Hare, Christian Thomas, Rolf Mathewes, Catherine La Farge, André L. Martel, Peter D. Heintzman, and Beth Shapiro Abstract: A partial skeleton of a bison was recovered during residential house construction in Whitehorse, Yukon, Canada. The specimen represents a young (estimated 6 year old) bison individual that died, was partially scavenged by carnivores, and subsequently buried by calcareous silt sediment in a pond or small lake during the middle Holocene, ϳ5400 years ago. Palaeoenvironmental data, including molluscs, pollen, vascular plant, and bryophyte macrofossils demonstrate that the small waterbody was surrounded by white spruce dominated boreal forest. Morphometric analysis of the skeleton reveals that its taxonomic affinity is ambiguous, likely owing to it representing an ontogenetically young individual, though it does share some cranial and horn core characteristics of named species such as Bison occidentalis or Bison priscus. Mitochondrial genomic data confirm that this bison belongs to Clade 2A (northern clade), which represents Pleistocene steppe bison (B. cf. priscus) in Beringia through the Holocene and is not represented in living bison species. These data further demonstrate that northern steppe bison population survived the late Pleistocene extinction event, persisted locally in southern Yukon into the Holocene, and are best characterized as a species with a high degree of morphological variability and ecological flexibility. Résumé : Le squelette partiel d’un bison a été récupéré durant la construction d’immeubles résidentiels a` Whitehorse (Yukon, Canada). Le spécimen représente un unique jeune bison (d’âge estimé a` 6 ans) qui est mort, a été partiellement dévoré par des carnivores pour ensuite être enseveli sous du silt calcareux dans un étang ou un petit lac durant l’Holocène moyen, il y a ϳ5400 ans. -
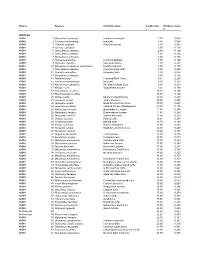
PDF File Containing Table of Lengths and Thicknesses of Turtle Shells And
Source Species Common name length (cm) thickness (cm) L t TURTLES AMNH 1 Sternotherus odoratus common musk turtle 2.30 0.089 AMNH 2 Clemmys muhlenbergi bug turtle 3.80 0.069 AMNH 3 Chersina angulata Angulate tortoise 3.90 0.050 AMNH 4 Testudo carbonera 6.97 0.130 AMNH 5 Sternotherus oderatus 6.99 0.160 AMNH 6 Sternotherus oderatus 7.00 0.165 AMNH 7 Sternotherus oderatus 7.00 0.165 AMNH 8 Homopus areolatus Common padloper 7.95 0.100 AMNH 9 Homopus signatus Speckled tortoise 7.98 0.231 AMNH 10 Kinosternon subrabum steinochneri Florida mud turtle 8.90 0.178 AMNH 11 Sternotherus oderatus Common musk turtle 8.98 0.290 AMNH 12 Chelydra serpentina Snapping turtle 8.98 0.076 AMNH 13 Sternotherus oderatus 9.00 0.168 AMNH 14 Hardella thurgi Crowned River Turtle 9.04 0.263 AMNH 15 Clemmys muhlenbergii Bog turtle 9.09 0.231 AMNH 16 Kinosternon subrubrum The Eastern Mud Turtle 9.10 0.253 AMNH 17 Kinixys crosa hinged-back tortoise 9.34 0.160 AMNH 18 Peamobates oculifers 10.17 0.140 AMNH 19 Peammobates oculifera 10.27 0.140 AMNH 20 Kinixys spekii Speke's hinged tortoise 10.30 0.201 AMNH 21 Terrapene ornata ornate box turtle 10.30 0.406 AMNH 22 Terrapene ornata North American box turtle 10.76 0.257 AMNH 23 Geochelone radiata radiated tortoise (Madagascar) 10.80 0.155 AMNH 24 Malaclemys terrapin diamondback terrapin 11.40 0.295 AMNH 25 Malaclemys terrapin Diamondback terrapin 11.58 0.264 AMNH 26 Terrapene carolina eastern box turtle 11.80 0.259 AMNH 27 Chrysemys picta Painted turtle 12.21 0.267 AMNH 28 Chrysemys picta painted turtle 12.70 0.168 AMNH 29 -

European Bison
IUCN/Species Survival Commission Status Survey and Conservation Action Plan The Species Survival Commission (SSC) is one of six volunteer commissions of IUCN – The World Conservation Union, a union of sovereign states, government agencies and non- governmental organisations. IUCN has three basic conservation objectives: to secure the conservation of nature, and especially of biological diversity, as an essential foundation for the future; to ensure that where the Earth’s natural resources are used this is done in a wise, European Bison equitable and sustainable way; and to guide the development of human communities towards ways of life that are both of good quality and in enduring harmony with other components of the biosphere. A volunteer network comprised of some 8,000 scientists, field researchers, government officials Edited by Zdzis³aw Pucek and conservation leaders from nearly every country of the world, the SSC membership is an Compiled by Zdzis³aw Pucek, Irina P. Belousova, unmatched source of information about biological diversity and its conservation. As such, SSC Ma³gorzata Krasiñska, Zbigniew A. Krasiñski and Wanda Olech members provide technical and scientific counsel for conservation projects throughout the world and serve as resources to governments, international conventions and conservation organisations. IUCN/SSC Action Plans assess the conservation status of species and their habitats, and specifies conservation priorities. The series is one of the world’s most authoritative sources of species conservation information -

Aurochs Genetics, a Cornerstone of European Biodiversity
Aurochs genetics, a cornerstone of European biodiversity Picture: Manolo Uno (c) Staffan Widstrand Authors: • drs. Ronald Goderie (Taurus Foundation); • dr. Johannes A. Lenstra (Utrecht University, Faculty of Veterinary Medicine); • Maulik Upadhyay (pHD Wageningen University); • dr. Richard Crooijmans (Animal Breeding and Genomics Centre, Wageningen University); • ir. Leo Linnartz (Ark Nature) Summary of: Aurochs Genetics, a cornerstone of biodiversity Preface In 2015 a report is written on Aurochs genetics, made possible by a grant from the Dutch Liberty Wildlife fund. This fund provided the Taurus foundation with a grant of EUR 20.000 to conduct genetic research on aurochs and its relation with nowadays so- called ‘primitive’ breeds. This is the summary of that report. This summary shortly describes the current state of affairs, what we do know early 2015 about the aurochs, about domestic cattle and the relationship of aurochs and the primitive breeds used in the Tauros Programme. Nijmegen, December 2015. page 2 Summary of: Aurochs Genetics, a cornerstone of biodiversity Table of contents Preface 2 Table of contents ......................................................................................................... 3 Summary ..................................................................................................................... 4 1 Introduction .......................................................................................................... 6 2 Aurochs: a short description ................................................................................. -

Tiger in Iran
ISSN 1027-2992 I Special Issue I N° 10 | Autumn 2016 CatsCAT in Iran news 02 CATnews is the newsletter of the Cat Specialist Group, a component Editors: Christine & Urs Breitenmoser of the Species Survival Commission SSC of the International Union Co-chairs IUCN/SSC for Conservation of Nature (IUCN). It is published twice a year, and is Cat Specialist Group available to members and the Friends of the Cat Group. KORA, Thunstrasse 31, 3074 Muri, Switzerland For joining the Friends of the Cat Group please contact Tel ++41(31) 951 90 20 Christine Breitenmoser at [email protected] Fax ++41(31) 951 90 40 <[email protected]> Original contributions and short notes about wild cats are welcome Send <[email protected]> contributions and observations to [email protected]. Guidelines for authors are available at www.catsg.org/catnews Cover Photo: From top left to bottom right: Caspian tiger (K. Rudloff) This Special Issue of CATnews has been produced with support Asiatic lion (P. Meier) from the Wild Cat Club and Zoo Leipzig. Asiatic cheetah (ICS/DoE/CACP/ Panthera) Design: barbara surber, werk’sdesign gmbh caracal (M. Eslami Dehkordi) Layout: Christine Breitenmoser & Tabea Lanz Eurasian lynx (F. Heidari) Print: Stämpfli Publikationen AG, Bern, Switzerland Pallas’s cat (F. Esfandiari) Persian leopard (S. B. Mousavi) ISSN 1027-2992 © IUCN/SSC Cat Specialist Group Asiatic wildcat (S. B. Mousavi) sand cat (M. R. Besmeli) jungle cat (B. Farahanchi) The designation of the geographical entities in this publication, and the representation of the material, do not imply the expression of any opinion whatsoever on the part of the IUCN concerning the legal status of any country, territory, or area, or its authorities, or concerning the delimitation of its frontiers or boundaries. -

A Middle Holocene Steppe Bison and Paleoenvironments from the Versleuce Meadows, Whitehorse, Yukon, Canada Grant D
1138 ARTICLE A middle Holocene steppe bison and paleoenvironments from the Versleuce Meadows, Whitehorse, Yukon, Canada Grant D. Zazula, Elizabeth Hall, P. Gregory Hare, Christian Thomas, Rolf Mathewes, Catherine La Farge, André L. Martel, Peter D. Heintzman, and Beth Shapiro Abstract: A partial skeleton of a bison was recovered during residential house construction in Whitehorse, Yukon, Canada. The specimen represents a young (estimated 6 year old) bison individual that died, was partially scavenged by carnivores, and subsequently buried by calcareous silt sediment in a pond or small lake during the middle Holocene, ϳ5400 years ago. Palaeoenvironmental data, including molluscs, pollen, vascular plant, and bryophyte macrofossils demonstrate that the small waterbody was surrounded by white spruce dominated boreal forest. Morphometric analysis of the skeleton reveals that its taxonomic affinity is ambiguous, likely owing to it representing an ontogenetically young individual, though it does share some cranial and horn core characteristics of named species such as Bison occidentalis or Bison priscus. Mitochondrial genomic data confirm that this bison belongs to Clade 2A (northern clade), which represents Pleistocene steppe bison (B. cf. priscus) in Beringia through the Holocene and is not represented in living bison species. These data further demonstrate that northern steppe bison population survived the late Pleistocene extinction event, persisted locally in southern Yukon into the Holocene, and are best characterized as a species with a high degree of morphological variability and ecological flexibility. Résumé : Le squelette partiel d’un bison a été récupéré durant la construction d’immeubles résidentiels a` Whitehorse (Yukon, Canada). Le spécimen représente un unique jeune bison (d’âge estimé a` 6 ans) qui est mort, a été partiellement dévoré par des carnivores pour ensuite être enseveli sous du silt calcareux dans un étang ou un petit lac durant l’Holocène moyen, il y a ϳ5400 ans. -

Bison Rewilding Plan 2014–2024 Rewilding Europe’S Contribution to the Comeback of the European Bison
Bison Rewilding Plan 2014–2024 Rewilding Europe’s contribution to the comeback of the European bison Advised by the Zoological Society of London Rewilding Europe This report was made possible by generous grants Bison Rewilding Plan, 2014–2024 by the Swedish Postcode Lottery (Sweden) and the Liberty Wildlife Fund (The Netherlands). Author Joep van de Vlasakker, Rewilding Europe Advised by Dr Jennifer Crees, Zoological Society of London Dr Monika Böhm, Zoological Society of London Peer-reviewed by Prof Jens-Christian Svenning, Aarhus University, Denmark Dr Rafal Kowalczyk, Director Mammal Research Institute / Polish Academy of Sciences / Bialowieza A report by Rewilding Europe Toernooiveld 1 6525 ED Nijmegen The Netherlands www.rewildingeurope.com About Rewilding Europe Founded in 2011, Rewilding Europe (RE) wants to make Europe a wilder place, with much more space for wildlife, wilderness and natural processes, by bringing back a variety of wildlife for all to enjoy and exploring new ways for people to earn a fair living from the wild. RE aims to rewild one million hectares of land by 2022, creating 10 magnificent wildlife and wilderness areas, which together reflect a wide selection of European regions and ecosystems, flora and fauna. Further information: www.rewildingeurope.com About ZSL Founded in 1826, the Zoological Society of London (ZSL) is an international scientific, conservation and educational charity whose vision is a world where animals are valued, and their conservation assured. Our mission, to promote and achieve the worldwide conservation of animals and their habitats, is realised through our groundbreaking science, our active conservation projects in more than 50 countries and our two Zoos, ZSL London Zoo and ZSL Whipsnade Zoo. -

Phylogeny of Dictyocaulus (Lungworms) from Eight Species of Ruminants Based on Analyses of Ribosomal RNA Data
179 Phylogeny of Dictyocaulus (lungworms) from eight species of ruminants based on analyses of ribosomal RNA data J. HO¨ GLUND1*,D.A.MORRISON1, B. P. DIVINA1,2, E. WILHELMSSON1 and J. G. MATTSSON1 1 Department of Parasitology (SWEPAR), National Veterinary Institute and Swedish University of Agriculture Sciences, 751 89 Uppsala, Sweden 2 College of Veterinary Medicine, University of the Philippines Los Ban˜os, College, Laguna, 4031 Philippines (Received 8 November 2002; revised 8 February 2003; accepted 8 February 2003) SUMMARY In this study, we conducted phylogenetic analyses of nematode parasites within the genus Dictyocaulus (superfamily Trichostrongyloidea). Lungworms from cattle (Bos taurus), domestic sheep (Ovis aries), European fallow deer (Dama dama), moose (Alces alces), musk ox (Ovibos moschatus), red deer (Cervus elaphus), reindeer (Rangifer tarandus) and roe deer (Capreolus capreolus) were obtained and their small subunit ribosomal RNA (SSU) and internal transcribed spacer 2 (ITS2) sequences analysed. In the hosts examined we identified D. capreolus, D. eckerti, D. filaria and D. viviparus. However, in fallow deer we detected a taxon with unique SSU and ITS2 sequences. The phylogenetic position of this taxon based on the SSU sequences shows that it is a separate evolutionary lineage from the other recognized species of Dictyocaulus. Furthermore, the analysis of the ITS2 sequence data indicates that it is as genetically distinct as are the named species of Dictyocaulus. Therefore, either this taxon needs to be recognized as a new species, or D. capreolus, D. eckerti and D. viviparus need to be combined into a single species. Traditionally, the genus Dictyocaulus has been placed as a separate family within the superfamily Trichostrongyloidea. -

Mixed-Species Exhibits with Pigs (Suidae)
Mixed-species exhibits with Pigs (Suidae) Written by KRISZTIÁN SVÁBIK Team Leader, Toni’s Zoo, Rothenburg, Luzern, Switzerland Email: [email protected] 9th May 2021 Cover photo © Krisztián Svábik Mixed-species exhibits with Pigs (Suidae) 1 CONTENTS INTRODUCTION ........................................................................................................... 3 Use of space and enclosure furnishings ................................................................... 3 Feeding ..................................................................................................................... 3 Breeding ................................................................................................................... 4 Choice of species and individuals ............................................................................ 4 List of mixed-species exhibits involving Suids ........................................................ 5 LIST OF SPECIES COMBINATIONS – SUIDAE .......................................................... 6 Sulawesi Babirusa, Babyrousa celebensis ...............................................................7 Common Warthog, Phacochoerus africanus ......................................................... 8 Giant Forest Hog, Hylochoerus meinertzhageni ..................................................10 Bushpig, Potamochoerus larvatus ........................................................................ 11 Red River Hog, Potamochoerus porcus ...............................................................