Host Specificity of Enterocytozoon Bieneusi Genotypes in Bactrian
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genotyping Approach for Potential Common Source of Enterocytozoon
SYNOPSIS Genotyping Approach for Potential Common Source of Enterocytozoon bieneusi Infection in Hematology Unit Guillaume Desoubeaux, Céline Nourrisson, Maxime Moniot, Marie-Alix De Kyvon, Virginie Bonnin, Marjan Ertault De La Bretonniére, Virginie Morange, Éric Bailly, Adrien Lemaignen, Florent Morio, Philippe Poirier Microsporidiosis is a fungal infection that generally causes wide range of host species (1,4). At least 16 microsporidian digestive disorders, especially in immunocompromised species have been described in humans, but Enterocytozoon hosts. Over a 4-day period in January 2018, 3 patients with bieneusi is the most common (5). However, little is known hematologic malignancies who were admitted to the hema- about the actual epidemiology of E. bieneusi microsporidi- tology unit of a hospital in France received diagnoses of En- osis, and there is a need for a better understanding of its terocytozoon bieneusi microsporidiosis. This unusually high pathophysiology and parasitic cycle (3,5). Unfortunately, incidence was investigated by sequence analysis at the in- ternal transcribed spacer rDNA locus and then by 3 micro- epidemiologic studies are complicated because E. bieneusi satellites and 1 minisatellite for multilocus genotyping. The infection has a low incidence rate worldwide (6), and its 3 isolates had many sequence similarities and belonged to microbiological diagnosis is difficult and likely often over- a new genotype closely related to genotype C. In addition, looked (7). In addition, the species is not easy to cultivate multilocus genotyping showed high genetic distances with in vitro in routine practice. Investigations can be carried out all the other strains collected from epidemiologically unre- directly only from infected biologic samples, which usually lated persons; none of these strains belonged to the new use DNA from fecal specimens (1,3,7). -

Bison Literature Review Biology
Bison Literature Review Ben Baldwin and Kody Menghini The purpose of this document is to compare the biology, ecology and basic behavior of cattle and bison for a management context. The literature related to bison is extensive and broad in scope covering the full continuum of domestication. The information incorporated in this review is focused on bison in more or less “wild” or free-ranging situations i.e.., not bison in close confinement or commercial production. While the scientific literature provides a solid basis for much of the basic biology and ecology, there is a wealth of information related to management implications and guidelines that is not captured. Much of the current information related to bison management, behavior (especially social organization) and practical knowledge is available through local experts, current research that has yet to be published, or popular literature. These sources, while harder to find and usually more localized in scope, provide crucial information pertaining to bison management. Biology Diet Composition Bison evolutional history provides the basis for many of the differences between bison and cattle. Bison due to their evolution in North America ecosystems are better adapted than introduced cattle, especially in grass dominated systems such as prairies. Many of these areas historically had relatively low quality forage. Bison are capable of more efficient digestion of low-quality forage then cattle (Peden et al. 1973; Plumb and Dodd 1993). Peden et al. (1973) also found that bison could consume greater quantities of low protein and poor quality forage then cattle. Bison and cattle have significant dietary overlap, but there are slight differences as well. -

Last Interglacial (MIS 5) Ungulate Assemblage from the Central Iberian Peninsula: the Camino Cave (Pinilla Del Valle, Madrid, Spain)
Palaeogeography, Palaeoclimatology, Palaeoecology 374 (2013) 327–337 Contents lists available at SciVerse ScienceDirect Palaeogeography, Palaeoclimatology, Palaeoecology journal homepage: www.elsevier.com/locate/palaeo Last Interglacial (MIS 5) ungulate assemblage from the Central Iberian Peninsula: The Camino Cave (Pinilla del Valle, Madrid, Spain) Diego J. Álvarez-Lao a,⁎, Juan L. Arsuaga b,c, Enrique Baquedano d, Alfredo Pérez-González e a Área de Paleontología, Departamento de Geología, Universidad de Oviedo, C/Jesús Arias de Velasco, s/n, 33005 Oviedo, Spain b Centro Mixto UCM-ISCIII de Evolución y Comportamiento Humanos, C/Sinesio Delgado, 4, 28029 Madrid, Spain c Departamento de Paleontología, Facultad de Ciencias Geológicas, Universidad Complutense de Madrid, Ciudad Universitaria, 28040 Madrid, Spain d Museo Arqueológico Regional de la Comunidad de Madrid, Plaza de las Bernardas, s/n, 28801-Alcalá de Henares, Madrid, Spain e Centro Nacional de Investigación sobre la Evolución Humana (CENIEH), Paseo Sierra de Atapuerca, s/n, 09002 Burgos, Spain article info abstract Article history: The fossil assemblage from the Camino Cave, corresponding to the late MIS 5, constitutes a key record to un- Received 2 November 2012 derstand the faunal composition of Central Iberia during the last Interglacial. Moreover, the largest Iberian Received in revised form 21 January 2013 fallow deer fossil population was recovered here. Other ungulate species present at this assemblage include Accepted 31 January 2013 red deer, roe deer, aurochs, chamois, wild boar, horse and steppe rhinoceros; carnivores and Neanderthals Available online 13 February 2013 are also present. The origin of the accumulation has been interpreted as a hyena den. Abundant fallow deer skeletal elements allowed to statistically compare the Camino Cave fossils with other Keywords: Early Late Pleistocene Pleistocene and Holocene European populations. -

Alternatives in Molecular Diagnostics of Encephalitozoon and Enterocytozoon Infections
Journal of Fungi Review Alternatives in Molecular Diagnostics of Encephalitozoon and Enterocytozoon Infections Alexandra Valenˇcáková * and Monika Suˇcik Department of Biology and Genetics, University of Veterinary Medicine and Pharmacy, Komenského 73, 04181 Košice, Slovakia; [email protected] * Correspondence: [email protected] Received: 15 June 2020; Accepted: 20 July 2020; Published: 22 July 2020 Abstract: Microsporidia are obligate intracellular pathogens that are currently considered to be most directly aligned with fungi. These fungal-related microbes cause infections in every major group of animals, both vertebrate and invertebrate, and more recently, because of AIDS, they have been identified as significant opportunistic parasites in man. The Microsporidia are ubiquitous parasites in the animal kingdom but, until recently, they have maintained relative anonymity because of the specialized nature of pathology researchers. Diagnosis of microsporidia infection from stool examination is possible and has replaced biopsy as the initial diagnostic procedure in many laboratories. These staining techniques can be difficult, however, due to the small size of the spores. The specific identification of microsporidian species has classically depended on ultrastructural examination. With the cloning of the rRNA genes from the human pathogenic microsporidia it has been possible to apply polymerase chain reaction (PCR) techniques for the diagnosis of microsporidial infection at the species and genotype level. The absence of genetic techniques for manipulating microsporidia and their complicated diagnosis hampered research. This study should provide basic insights into the development of diagnostics and the pitfalls of molecular identification of these ubiquitous intracellular pathogens that can be integrated into studies aimed at treating or controlling microsporidiosis. Keywords: Encephalitozoon spp.; Enterocytozoonbieneusi; diagnosis; molecular diagnosis; primers 1. -
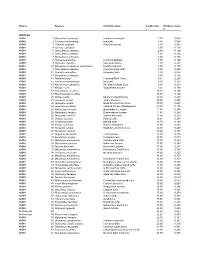
PDF File Containing Table of Lengths and Thicknesses of Turtle Shells And
Source Species Common name length (cm) thickness (cm) L t TURTLES AMNH 1 Sternotherus odoratus common musk turtle 2.30 0.089 AMNH 2 Clemmys muhlenbergi bug turtle 3.80 0.069 AMNH 3 Chersina angulata Angulate tortoise 3.90 0.050 AMNH 4 Testudo carbonera 6.97 0.130 AMNH 5 Sternotherus oderatus 6.99 0.160 AMNH 6 Sternotherus oderatus 7.00 0.165 AMNH 7 Sternotherus oderatus 7.00 0.165 AMNH 8 Homopus areolatus Common padloper 7.95 0.100 AMNH 9 Homopus signatus Speckled tortoise 7.98 0.231 AMNH 10 Kinosternon subrabum steinochneri Florida mud turtle 8.90 0.178 AMNH 11 Sternotherus oderatus Common musk turtle 8.98 0.290 AMNH 12 Chelydra serpentina Snapping turtle 8.98 0.076 AMNH 13 Sternotherus oderatus 9.00 0.168 AMNH 14 Hardella thurgi Crowned River Turtle 9.04 0.263 AMNH 15 Clemmys muhlenbergii Bog turtle 9.09 0.231 AMNH 16 Kinosternon subrubrum The Eastern Mud Turtle 9.10 0.253 AMNH 17 Kinixys crosa hinged-back tortoise 9.34 0.160 AMNH 18 Peamobates oculifers 10.17 0.140 AMNH 19 Peammobates oculifera 10.27 0.140 AMNH 20 Kinixys spekii Speke's hinged tortoise 10.30 0.201 AMNH 21 Terrapene ornata ornate box turtle 10.30 0.406 AMNH 22 Terrapene ornata North American box turtle 10.76 0.257 AMNH 23 Geochelone radiata radiated tortoise (Madagascar) 10.80 0.155 AMNH 24 Malaclemys terrapin diamondback terrapin 11.40 0.295 AMNH 25 Malaclemys terrapin Diamondback terrapin 11.58 0.264 AMNH 26 Terrapene carolina eastern box turtle 11.80 0.259 AMNH 27 Chrysemys picta Painted turtle 12.21 0.267 AMNH 28 Chrysemys picta painted turtle 12.70 0.168 AMNH 29 -

European Bison
IUCN/Species Survival Commission Status Survey and Conservation Action Plan The Species Survival Commission (SSC) is one of six volunteer commissions of IUCN – The World Conservation Union, a union of sovereign states, government agencies and non- governmental organisations. IUCN has three basic conservation objectives: to secure the conservation of nature, and especially of biological diversity, as an essential foundation for the future; to ensure that where the Earth’s natural resources are used this is done in a wise, European Bison equitable and sustainable way; and to guide the development of human communities towards ways of life that are both of good quality and in enduring harmony with other components of the biosphere. A volunteer network comprised of some 8,000 scientists, field researchers, government officials Edited by Zdzis³aw Pucek and conservation leaders from nearly every country of the world, the SSC membership is an Compiled by Zdzis³aw Pucek, Irina P. Belousova, unmatched source of information about biological diversity and its conservation. As such, SSC Ma³gorzata Krasiñska, Zbigniew A. Krasiñski and Wanda Olech members provide technical and scientific counsel for conservation projects throughout the world and serve as resources to governments, international conventions and conservation organisations. IUCN/SSC Action Plans assess the conservation status of species and their habitats, and specifies conservation priorities. The series is one of the world’s most authoritative sources of species conservation information -

Supplemental Figure 2.Pdf
Supplemental Figure 2. Published genomes used in this study Genome code proteome_file Genus species Acain1_GeneCatalog_proteins_20150 Acain1 309.aa.fasta Acaromyces ingoldii MCA 4198 Aciri1_iso_GeneCatalog_proteins_20 Aciri1iso 111207.aa.fasta Acidomyces richmondensis Agabivarbi Abisporus_varbisporusH97.v2.Filtere Agaricus bisporus var bisporus sH972 dModels3.proteins.fasta H97 Agabivarb Abisporus_varburnettii.v2.FilteredMo Agaricus bisporus var. burnettii ur1 dels1.proteins.fasta JB137-S8 Altal1_GeneCatalog_proteins_201410 Altal1 10.aa.fasta Alternaria alternata SRC1lrK2f Amamu1_GeneCatalog_proteins_201 Amamu1 20806.aa.fasta Amanita muscaria Koide Amath1_GeneCatalog_proteins_2012 Amath1 0702.aa.fasta Amanita thiersii Skay4041 Amore1_GeneCatalog_proteins_2011 Amore1 0719.aa.fasta Amorphotheca resinae Antav1_GeneCatalog_proteins_20130 Anthostoma_avocetta_NRRL_31 Antav1 828.aa.fasta 90 Armme1_1_GeneCatalog_proteins_20 Armme11 140802.aa.fasta Armillaria mellea Armos1 Armos1.aa.fasta Armillaria ostoye Artol1_GeneCatalog_proteins_201312 Arthrobotrys oligospora ATCC Artol1 09.aa.fasta 24927 Artbe1_GeneCatalog_proteins_20131 Arthroderma benhamiae CBS Artbe1 209.aa.fasta 112371 Artel1_GeneCatalog_proteins_201406 Artel1 30.aa.fasta Artolenzites elegans CIRM1663 Ascim1_GeneCatalog_proteins_20121 Ascim1 221.aa.fasta Ascobolus immersus RN42 Ascsa1_GeneCatalog_proteins_20140 Ascocoryne_sarcoides_NRRL500 Ascsa1 116.aa.fasta 72 Ascru1_GeneCatalog_proteins_20111 Ascoidea rubescens NRRL Ascru1 107.aa.fasta Y17699 Ashgo1_GeneCatalog_proteins_20130 Ashgo1 -

Enterocytozoon Bieneusi and Encephalitozoon Spp
Deng et al. BMC Veterinary Research (2020) 16:212 https://doi.org/10.1186/s12917-020-02434-z RESEARCH ARTICLE Open Access First identification and genotyping of Enterocytozoon bieneusi and Encephalitozoon spp. in pet rabbits in China Lei Deng†, Yijun Chai†, Leiqiong Xiang†, Wuyou Wang†, Ziyao Zhou, Haifeng Liu, Zhijun Zhong, Hualin Fu and Guangneng Peng* Abstract Background: Microsporidia are common opportunistic parasites in humans and animals, including rabbits. However, only limited epidemiology data concern about the prevalence and molecular characterization of Enterocytozoon bieneusi and Encephalitozoon spp. in rabbits. This study is the first detection and genotyping of Microsporidia in pet rabbits in China. Results: A total of 584 faecal specimens were collected from rabbits in pet shops from four cities in Sichuan province, China. The overall prevalence of microsporidia infection was 24.8% by nested PCR targeting the internal transcribed spacer (ITS) region of E. bieneusi and Encephalitozoon spp. respectively. E. bieneusi was the most common species (n = 90, 15.4%), followed by Encephalitozoon cuniculi (n = 34, 5.8%) and Encephalitozoon intestinalis (n = 16, 2.7%). Mixed infections (E. bieneusi and E. cuniculi) were detected in five another rabbits (0.9%). Statistically significant differences in the prevalence of microsporidia were observed among different cities (χ2 = 38.376, df = 3, P < 0.01) and the rabbits older than 1 year were more likely to harbour microsporidia infections (χ2 =9.018,df=2,P <0.05).Elevendistinct genotypes of E. bieneusi were obtained, including five known (SC02, I, N, J, CHY1) and six novel genotypes (SCR01, SCR02, SCR04 to SCR07). SC02 was the most prevalent genotype in all tested cities (43.3%, 39/90). -

Species-Specific Identification of Microsporidia in Stool and Intestinal Biopsy Specimens by the Polymerase Chain Reaction
Article Vol. 16, No. 5 369 Eur. J. Clin. Microbiol. Infect. Dis., 1997, 16:369-376 Species-Specific Identification of Microsporidia in Stool and Intestinal Biopsy Specimens by the Polymerase Chain Reaction N.R Kock 1., H. Petersen 2, T. Fenner 2, I. Sobottka 3, C. Schmetz 4, R Deplazes 5, N.J. Pieniazek 6, H. Albrecht 7, J. Schottelius I In view of the increasing number of cases of human microsporidiosis, simple and rapid methods for clear identification of microsporidian parasites to the species level are re- quired. In the present study, the polymerase chain reaction (PCR) was used for species- specific detection of Encephalitozoon cunicufi, Encephalitozoon hellem, Encephafitozoon (Septata) intestinalis, and Enterocytozoon bieneusi in both tissue and stool. Using stool specimens and intestinal biopsies of patients infected with Enterocytozoon bieneusi (n = 9), Encephalitozoon spp. (n = 2), and Encephafitozoon intestinalis (n = 1) as well as stool spiked with spores of Encephafitozoon cunicufi and Encephalitozoon hellem and tissue cultures of Encephalitozoon cuniculi and Encephalitozoon hellem, three proce- dures were developed to produce PCR-ready DNA directly from the samples. Specific detection of microsporidian pathogens was achieved in the first PCR. The subsequent nested PCR permitted species determination and verified the first PCR products. With- out exception, the PCR assay confirmed electron microscopic detection of Enterocyto- zoon bieneusi and Encephalitozoon intestinalis in stool specimens and their correspond- ing biopsies and in spiked stool samples and tissue cultures infected with Encephalito- zoon cuniculi and Encephafitozoon hellem. Moreover, identification of Encephafito- zoon spp. could be specified as Encephalitozoon intestinalis. Whereas standard meth- ods such as light and transmission electron microscopy may lack sensitivity or require more time and special equipment, the PCR procedure described facilitates species- specific identification of microsporidian parasites in stool, biopsies, and, probably, other samples in about five hours. -

Phylogeny of Dictyocaulus (Lungworms) from Eight Species of Ruminants Based on Analyses of Ribosomal RNA Data
179 Phylogeny of Dictyocaulus (lungworms) from eight species of ruminants based on analyses of ribosomal RNA data J. HO¨ GLUND1*,D.A.MORRISON1, B. P. DIVINA1,2, E. WILHELMSSON1 and J. G. MATTSSON1 1 Department of Parasitology (SWEPAR), National Veterinary Institute and Swedish University of Agriculture Sciences, 751 89 Uppsala, Sweden 2 College of Veterinary Medicine, University of the Philippines Los Ban˜os, College, Laguna, 4031 Philippines (Received 8 November 2002; revised 8 February 2003; accepted 8 February 2003) SUMMARY In this study, we conducted phylogenetic analyses of nematode parasites within the genus Dictyocaulus (superfamily Trichostrongyloidea). Lungworms from cattle (Bos taurus), domestic sheep (Ovis aries), European fallow deer (Dama dama), moose (Alces alces), musk ox (Ovibos moschatus), red deer (Cervus elaphus), reindeer (Rangifer tarandus) and roe deer (Capreolus capreolus) were obtained and their small subunit ribosomal RNA (SSU) and internal transcribed spacer 2 (ITS2) sequences analysed. In the hosts examined we identified D. capreolus, D. eckerti, D. filaria and D. viviparus. However, in fallow deer we detected a taxon with unique SSU and ITS2 sequences. The phylogenetic position of this taxon based on the SSU sequences shows that it is a separate evolutionary lineage from the other recognized species of Dictyocaulus. Furthermore, the analysis of the ITS2 sequence data indicates that it is as genetically distinct as are the named species of Dictyocaulus. Therefore, either this taxon needs to be recognized as a new species, or D. capreolus, D. eckerti and D. viviparus need to be combined into a single species. Traditionally, the genus Dictyocaulus has been placed as a separate family within the superfamily Trichostrongyloidea. -

Microsporidia: a Journey Through Radical Taxonomical Revisions
fungal biology reviews 23 (2009) 1–8 journal homepage: www.elsevier.com/locate/fbr Review Microsporidia: a journey through radical taxonomical revisions Nicolas CORRADI, Patrick J. KEELING* Canadian Institute for Advanced Research, Department of Botany, University of British Columbia, 3529-6270 University Boulevard, Vancouver, BC V6T 1Z4, Canada article info abstract Article history: Microsporidia are obligate intracellular parasites of medical and commercial importance, Received 3 March 2009 characterized by a severe reduction, or even absence, of cellular components typical of Received in revised form eukaryotes such as mitochondria, Golgi apparatus and flagella. This simplistic cellular 23 May 2009 organization has made it difficult to infer the evolutionary relationship of Microsporidia Accepted 28 May 2009 to other eukaryotes, because they lack many characters historically used to make such comparisons. Eventually, it was suggested that this simplicity might be due to Microspor- Keywords: idia representing a very early eukaryotic lineage that evolved prior to the origin of many Complexity typically eukaryotic features, in particular the mitochondrion. This hypothesis was sup- Genome reduction ported by the first biochemical and molecular studies of the group. In the last decade, Microsporidia however, contrasting evidence has emerged, mostly from molecular sequences, that Phylogeny show Microsporidia are related to fungi, and it is now widely acknowledged that features Simplicity previously recognized as primitive are instead highly derived adaptations to their obligate Taxonomy parasitic lifestyle. The various sharply differing views on microsporidian evolution resulted in several radical reappraisals of their taxonomy. Here we will chronologically review the causes and consequences for these taxonomic revisions, with a special emphasis on why the unique cellular and genomic features of Microsporidia lured scientists towards the wrong direction for so long. -

Evolution and Functional Insights of Different Ancestral Orthologous Clades of Chitin Synthase Genes in the Fungal Tree of Life
ORIGINAL RESEARCH published: 01 February 2016 doi: 10.3389/fpls.2016.00037 Evolution and Functional Insights of Different Ancestral Orthologous Clades of Chitin Synthase Genes in the Fungal Tree of Life Mu Li 1 †, Cong Jiang 1 †, Qinhu Wang 1, Zhongtao Zhao 2, Qiaojun Jin 1, Jin-Rong Xu 1, 3 and Huiquan Liu 1* Edited by: Thiago Motta Venancio, 1 State Key Laboratory of Crop Stress Biology for Arid Areas, College of Plant Protection, Northwest A&F University, Yangling, Universidade Estadual do Norte China, 2 South China Botanical Garden, Chinese Academy of Sciences, Guangzhou, China, 3 Department of Botany and Fluminense, Brazil Plant Pathology, Purdue University, West Lafayette, IN, USA Reviewed by: Lakshminarayan M. Iyer, Chitin synthases (CHSs) are key enzymes in the biosynthesis of chitin, an important National Center for Biotechnology Information, National Library of structural component of fungal cell walls that can trigger innate immune responses in Medicine, National Institutes of Health, host plants and animals. Members of CHS gene family perform various functions in fungal USA Robson Francisco De Souza, cellular processes. Previous studies focused primarily on classifying diverse CHSs into Universidade de São Paulo, Brazil different classes, regardless of their functional diversification, or on characterizing their A. Maxwell Burroughs, functions in individual fungal species. A complete and systematic comparative analysis National Center for Biotechnology Information, National Library of of CHS genes based on their orthologous relationships will be valuable for elucidating Medicine, National Institutes of Health, the evolution and functions of different CHS genes in fungi. Here, we identified and USA compared members of the CHS gene family across the fungal tree of life, including *Correspondence: Huiquan Liu 18 divergent fungal lineages.