Gene Name Description Studies Studies with FC Total Sample Sizes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

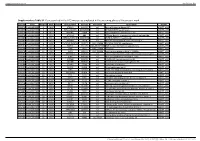
Table 2. Significant
Table 2. Significant (Q < 0.05 and |d | > 0.5) transcripts from the meta-analysis Gene Chr Mb Gene Name Affy ProbeSet cDNA_IDs d HAP/LAP d HAP/LAP d d IS Average d Ztest P values Q-value Symbol ID (study #5) 1 2 STS B2m 2 122 beta-2 microglobulin 1452428_a_at AI848245 1.75334941 4 3.2 4 3.2316485 1.07398E-09 5.69E-08 Man2b1 8 84.4 mannosidase 2, alpha B1 1416340_a_at H4049B01 3.75722111 3.87309653 2.1 1.6 2.84852656 5.32443E-07 1.58E-05 1110032A03Rik 9 50.9 RIKEN cDNA 1110032A03 gene 1417211_a_at H4035E05 4 1.66015788 4 1.7 2.82772795 2.94266E-05 0.000527 NA 9 48.5 --- 1456111_at 3.43701477 1.85785922 4 2 2.8237185 9.97969E-08 3.48E-06 Scn4b 9 45.3 Sodium channel, type IV, beta 1434008_at AI844796 3.79536664 1.63774235 3.3 2.3 2.75319499 1.48057E-08 6.21E-07 polypeptide Gadd45gip1 8 84.1 RIKEN cDNA 2310040G17 gene 1417619_at 4 3.38875643 1.4 2 2.69163229 8.84279E-06 0.0001904 BC056474 15 12.1 Mus musculus cDNA clone 1424117_at H3030A06 3.95752801 2.42838452 1.9 2.2 2.62132809 1.3344E-08 5.66E-07 MGC:67360 IMAGE:6823629, complete cds NA 4 153 guanine nucleotide binding protein, 1454696_at -3.46081884 -4 -1.3 -1.6 -2.6026947 8.58458E-05 0.0012617 beta 1 Gnb1 4 153 guanine nucleotide binding protein, 1417432_a_at H3094D02 -3.13334396 -4 -1.6 -1.7 -2.5946297 1.04542E-05 0.0002202 beta 1 Gadd45gip1 8 84.1 RAD23a homolog (S. -

Datasheet: VPA00586KT Product Details
Datasheet: VPA00586KT Description: EIF3E ANTIBODY WITH CONTROL LYSATE Specificity: EIF3E Format: Purified Product Type: PrecisionAb™ Polyclonal Isotype: Polyclonal IgG Quantity: 2 Westerns Product Details Applications This product has been reported to work in the following applications. This information is derived from testing within our laboratories, peer-reviewed publications or personal communications from the originators. Please refer to references indicated for further information. For general protocol recommendations, please visit www.bio-rad-antibodies.com/protocols. Yes No Not Determined Suggested Dilution Western Blotting 1/1000 PrecisionAb antibodies have been extensively validated for the western blot application. The antibody has been validated at the suggested dilution. Where this product has not been tested for use in a particular technique this does not necessarily exclude its use in such procedures. Further optimization may be required dependant on sample type. Target Species Human Species Cross Reacts with: Rat Reactivity N.B. Antibody reactivity and working conditions may vary between species. Product Form Purified IgG - liquid Preparation 20μl Rabbit polyclonal antibody purified by affinity chromatography Buffer Solution Phosphate buffered saline Preservative 0.09% Sodium Azide (NaN3) Stabilisers 2% Sucrose Immunogen Synthetic peptide directed towards the middle region of human EIF3E External Database Links UniProt: P60228 Related reagents Entrez Gene: 3646 EIF3E Related reagents Synonyms EIF3S6, INT6 Page 1 of 2 Specificity Rabbit anti Human EIF3E antibody recognizes the eukaryotic translation initiation factor 3 subunit E, also known as eIF-3 p48, eukaryotic translation initiation factor 3 subunit 6, mammary tumor- associated protein INT6 or viral integration site protein INT-6 homolog. Rabbit anti Human EIF3E antibody detects a band of 48 kDa. -

Apoptotic Genes As Potential Markers of Metastatic Phenotype in Human Osteosarcoma Cell Lines
17-31 10/12/07 14:53 Page 17 INTERNATIONAL JOURNAL OF ONCOLOGY 32: 17-31, 2008 17 Apoptotic genes as potential markers of metastatic phenotype in human osteosarcoma cell lines CINZIA ZUCCHINI1, ANNA ROCCHI2, MARIA CRISTINA MANARA2, PAOLA DE SANCTIS1, CRISTINA CAPANNI3, MICHELE BIANCHINI1, PAOLO CARINCI1, KATIA SCOTLANDI2 and LUISA VALVASSORI1 1Dipartimento di Istologia, Embriologia e Biologia Applicata, Università di Bologna, Via Belmeloro 8, 40126 Bologna; 2Laboratorio di Ricerca Oncologica, Istituti Ortopedici Rizzoli; 3IGM-CNR, Unit of Bologna, c/o Istituti Ortopedici Rizzoli, Via di Barbiano 1/10, 40136 Bologna, Italy Received May 29, 2007; Accepted July 19, 2007 Abstract. Metastasis is the most frequent cause of death among malignant primitive bone tumor, usually developing in children patients with osteosarcoma. We have previously demonstrated and adolescents, with a high tendency to metastasize (2). in independent experiments that the forced expression of Metastases in osteosarcoma patients spread through peripheral L/B/K ALP and CD99 in U-2 OS osteosarcoma cell lines blood very early and colonize primarily the lung, and later markedly reduces the metastatic ability of these cancer cells. other skeleton districts (3). Since disseminated hidden micro- This behavior makes these cell lines a useful model to assess metastases are present in 80-90% of OS patients at the time the intersection of multiple and independent gene expression of diagnosis, the identification of markers of invasiveness signatures concerning the biological problem of dissemination. and metastasis forms a target of paramount importance in With the aim to characterize a common transcriptional profile planning the treatment of osteosarcoma lesions and enhancing reflecting the essential features of metastatic behavior, we the prognosis. -
Supplementary Table S1. Genes Printed in the HC5 Microarray Employed in the Screening Phase of the Present Work
Supplementary material Ann Rheum Dis Supplementary Table S1. Genes printed in the HC5 microarray employed in the screening phase of the present work. CloneID Plate Position Well Length GeneSymbol GeneID Accession Description Vector 692672 HsxXG013989 2 B01 STK32A 202374 null serine/threonine kinase 32A pANT7_cGST 692675 HsxXG013989 3 C01 RPS10-NUDT3 100529239 null RPS10-NUDT3 readthrough pANT7_cGST 692678 HsxXG013989 4 D01 SPATA6L 55064 null spermatogenesis associated 6-like pANT7_cGST 692679 HsxXG013989 5 E01 ATP1A4 480 null ATPase, Na+/K+ transporting, alpha 4 polypeptide pANT7_cGST 692689 HsxXG013989 6 F01 ZNF816-ZNF321P 100529240 null ZNF816-ZNF321P readthrough pANT7_cGST 692691 HsxXG013989 7 G01 NKAIN1 79570 null Na+/K+ transporting ATPase interacting 1 pANT7_cGST 693155 HsxXG013989 8 H01 TNFSF12-TNFSF13 407977 NM_172089 TNFSF12-TNFSF13 readthrough pANT7_cGST 693161 HsxXG013989 9 A02 RAB12 201475 NM_001025300 RAB12, member RAS oncogene family pANT7_cGST 693169 HsxXG013989 10 B02 SYN1 6853 NM_133499 synapsin I pANT7_cGST 693176 HsxXG013989 11 C02 GJD3 125111 NM_152219 gap junction protein, delta 3, 31.9kDa pANT7_cGST 693181 HsxXG013989 12 D02 CHCHD10 400916 null coiled-coil-helix-coiled-coil-helix domain containing 10 pANT7_cGST 693184 HsxXG013989 13 E02 IDNK 414328 null idnK, gluconokinase homolog (E. coli) pANT7_cGST 693187 HsxXG013989 14 F02 LYPD6B 130576 null LY6/PLAUR domain containing 6B pANT7_cGST 693189 HsxXG013989 15 G02 C8orf86 389649 null chromosome 8 open reading frame 86 pANT7_cGST 693194 HsxXG013989 16 H02 CENPQ 55166 -

EIF3E (NM 001568) Human Recombinant Protein Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TP304788 EIF3E (NM_001568) Human Recombinant Protein Product data: Product Type: Recombinant Proteins Description: Recombinant protein of human eukaryotic translation initiation factor 3, subunit E (EIF3E) Species: Human Expression Host: HEK293T Tag: C-Myc/DDK Predicted MW: 52 kDa Concentration: >50 ug/mL as determined by microplate BCA method Purity: > 80% as determined by SDS-PAGE and Coomassie blue staining Buffer: 25 mM Tris.HCl, pH 7.3, 100 mM glycine, 10% glycerol Preparation: Recombinant protein was captured through anti-DDK affinity column followed by conventional chromatography steps. Storage: Store at -80°C. Stability: Stable for 12 months from the date of receipt of the product under proper storage and handling conditions. Avoid repeated freeze-thaw cycles. RefSeq: NP_001559 Locus ID: 3646 UniProt ID: P60228 RefSeq Size: 1516 Cytogenetics: 8q23.1 RefSeq ORF: 1335 Synonyms: eIF3-p46; EIF3-P48; EIF3S6; INT6 This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 2 EIF3E (NM_001568) Human Recombinant Protein – TP304788 Summary: Component of the eukaryotic translation initiation factor 3 (eIF-3) complex, which is required for several steps in the initiation of protein synthesis (PubMed:17581632, PubMed:25849773, PubMed:27462815). The eIF-3 complex associates with the 40S ribosome and facilitates the recruitment of eIF-1, eIF-1A, eIF-2:GTP:methionyl-tRNAi and eIF-5 to form the 43S pre- initiation complex (43S PIC). -

Genename Sequence Peptidecount
GeneName Sequence PeptideCount A1BG SLPAPWLSMAPVSWITPGLK 1 A1S9T;UBA1;UBE1 AAVATFLQSVQVPEFTPK 18 A1S9T;UBA1;UBE1 AENYDIPSADR 18 A1S9T;UBA1;UBE1 ALPAVQQNNLDEDLIR 18 A1S9T;UBA1;UBE1 ALPAVQQNNLDEDLIRK 18 A1S9T;UBA1;UBE1 ATLPSPDKLPGFK 18 A1S9T;UBA1;UBE1 DEFEGLFK 18 A1S9T;UBA1;UBE1 GGIVSQVK 18 A1S9T;UBA1;UBE1 GLGVEIAK 18 A1S9T;UBA1;UBE1 HQYYNQEWTLWDR 18 A1S9T;UBA1;UBE1 IYDDDFFQNLDGVANALDNVDAR 18 A1S9T;UBA1;UBE1 KPLLESGTLGTK 18 A1S9T;UBA1;UBE1 LAGTQPLEVLEAVQR 18 A1S9T;UBA1;UBE1 LQTSSVLVSGLR 18 A1S9T;UBA1;UBE1 NEEDAAELVALAQAVNAR 18 A1S9T;UBA1;UBE1 NFPNAIEHTLQWAR 18 A1S9T;UBA1;UBE1 QFLDYFK 18 A1S9T;UBA1;UBE1 QFLFRPWDVTK 18 A1S9T;UBA1;UBE1 SLVASLAEPDFVVTDFAK 18 A2LD1 TLEPYPLVIAGEHNIPWLLHLPGSGR 1 A4;AD1;APP THPHFVIPYR 2 A4;AD1;APP WYFDVTEGK 2 AAAS;ADRACALA;GL003 GGGVTNLLWSPDGSK 12 AAAS;ADRACALA;GL003 GQWINLPVLQLTK 12 AAAS;ADRACALA;GL003 IAHIPLYFVNAQFPR 12 AAAS;ADRACALA;GL003 ILATTPSAVFR 12 AAAS;ADRACALA;GL003 LAVLMK 12 AAAS;ADRACALA;GL003 LLSASPVDAAIR 12 AAAS;ADRACALA;GL003 SATIVADLSETTIQTPDGEER 12 AAAS;ADRACALA;GL003 VFAWHPHTNK 12 AAAS;ADRACALA;GL003 VQDGKPVILLFR 12 AAAS;ADRACALA;GL003 VWEAQMWTCER 12 AAAS;ADRACALA;GL003 VYNASSTIVPSLK 12 AAAS;ADRACALA;GL003 WPTLSGR 12 AAC4;ANT4;SFEC;SLC25A31 DLLAGGVAAAVSK 21 AAC4;ANT4;SFEC;SLC25A31 EQGFFSFWR 21 AAC4;ANT4;SFEC;SLC25A31 GAFSNVLR 21 AAC4;ANT4;SFEC;SLC25A31 GLGDCIMK 21 AAC4;ANT4;SFEC;SLC25A31 GLLPKPK 21 AAC4;ANT4;SFEC;SLC25A31 GMVDCLVR 21 AAC4;ANT4;SFEC;SLC25A31 GNLANVIR 21 AAC4;ANT4;SFEC;SLC25A31 GTGGALVLVLYDK 21 AAC4;ANT4;SFEC;SLC25A31 IKEFFHIDIGGR 21 AAC4;ANT4;SFEC;SLC25A31 IPREQGFFSFWR 21 AAC4;ANT4;SFEC;SLC25A31 -

N'-1- Gene a in Colon Cancer Mrna - Sy?- Gene a in Lung Cancer Mrna
US 20080045471 A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2008/0045471 A1 NOrth (43) Pub. Date: Feb. 21, 2008 (54) NUCLEC ACDS FOR APOPTOSIS OF Publication Classification CANCER CELLS (51) Int. Cl. A6IR 48/00 (2006.01) (76) Inventor: Don Adams North, Arlington, TX (US) C7H 2L/00 (2006.01) CI2N 5/06 (2006.01) Correspondence Address: (52) U.S. Cl. ............................ 514/44; 435/375; 536/23.1 BAKER BOTTS LLP. (57) ABSTRACT PATENT DEPARTMENT The disclosure relates to nucleic acids having Apoptotic 98 SANJACINTO BLVD., SUITE 1500 Sequence Nos. 5, 8, 9, 11, 14, 60 and 66. It also relates to AUSTIN, TX 78701-4039 (US) agents targeting Apoptotic Sequences, said agents having SEQID NO:1, SEQID NO:2, SEQID NO:3, SEQID NO:4, SEQ ID NO: 5, SEQ ID NO:6, and SEQ ID NO:7. The (21) Appl. No.: 11/691,994 composition may also include a pharmaceutically acceptable carrier. The disclosure also includes a method of killing a Filed: Mar. 27, 2007 cancer cell by administering to a cancer cell a treatment (22) formulation including a nucleic acid having an Apoptotic Related U.S. Application Data Sequence targeting agent of: SEQID NO:1, SEQID NO:2. SEQ ID NO:3, SEQ ID NO:4, SEQ ID NO. 5, SEQ ID (60) Provisional application No. 60/786,316, filed on Mar. NO:6, and SEQID NO:7 and a pharmaceutically acceptable 27, 2006. Provisional application No. 60/820,577, carrier. The cancer cell may be located in a subject with filed on Jul. -

Vaccine-Increased Seq ID Unigene ID Uniprot ID Gene Names
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) J Immunother Cancer Vaccine-Increased Seq ID Unigene ID Uniprot ID Gene Names 1_HSPA1A_3303 Hs.274402 P0DMV8 HSPA1A HSP72 HSPA1 HSX70 100_AKAP17A_8227 Hs.522572 Q02040 AKAP17A CXYorf3 DXYS155E SFRS17A XE7 1000_H2AFY_9555 Hs.420272 O75367 H2AFY MACROH2A1 1001_ITPK1_3705 Hs.308122 Q13572 ITPK1 1002_PTPN11_5781 Hs.506852 Q06124 PTPN11 PTP2C SHPTP2 1003_EIF3J_8669 Hs.404056 O75822 EIF3J EIF3S1 PRO0391 1004_TRIP12_9320 Hs.591633 Q14669 TRIP12 KIAA0045 ULF 1006_YEATS2_55689 Hs.632575 Q9ULM3 YEATS2 KIAA1197 1007_SEL1L3_23231 Hs.479384 Q68CR1 SEL1L3 KIAA0746 1008_IDH1_3417 Hs.593422 O75874 IDH1 PICD 101_HSPH1_10808 Hs.36927 Q92598 HSPH1 HSP105 HSP110 KIAA0201 1010_LDLR_3949 Hs.213289 P01130 LDLR 1011_FAM129B_64855 Hs.522401 Q96TA1 NIBAN2 C9orf88 FAM129B 1012_MAP3K5_4217 Hs.186486 Q99683 MAP3K5 ASK1 MAPKKK5 MEKK5 1013_NEFH_4744 Hs.198760 P12036 NEFH KIAA0845 NFH 1014_RAP1B_5908 Hs.369920 P61224 RAP1B OK/SW-cl.11 1015_MCCC1_56922 Hs.47649 Q96RQ3 MCCC1 MCCA 1017_MT1E_4493 Hs.534330 P04732 MT1E 1022_TXNDC5_81567 Hs.150837 Q8NBS9 TXNDC5 TLP46 UNQ364/PRO700 1023_STRA13_201254 Hs.37616 O14503 BHLHE40 BHLHB2 DEC1 SHARP2 STRA13 1024_NPEPPS_9520 Hs.443837 P55786 NPEPPS PSA 1025_YIPF6_286451 Hs.82719 Q96EC8 YIPF6 1026_CLIP1_6249 Hs.524809 P30622 CLIP1 CYLN1 RSN 1027_SRSF7_6432 Hs.309090 Q16629 SRSF7 SFRS7 103_RPS25_6230 Hs.512676 P62851 RPS25 1031_SOCS7_30837 -

Original Article Overexpression of Eif3e Is Correlated with Colon Tumor Development and Poor Prognosis
Int J Clin Exp Pathol 2014;7(10):6462-6474 www.ijcep.com /ISSN:1936-2625/IJCEP0002023 Original Article Overexpression of eIF3e is correlated with colon tumor development and poor prognosis Zhi Li1*, Shengtao Lin2*, Tao Jiang3, Jingtao Wang2, Huijun Lu4, Huamei Tang4, Mujian Teng5, Junwei Fan2 1Department of Anorectal Surgery, Qianfoshan Hospital Affiliated to Shandong University, 16766 Jingshi Road, Jinan 250014, Shandong, China; 2Department of General Surgery, Shanghai Jiaotong University Affiliated First People’s Hospital, 85 Wujin Road, Shanghai 200080, China; 3Department of Anal-Colorectal Surgery, General Hospital of Ningxia Medical University, 804 South Shengli Road, Yinchuan 750004, China; 4Department of Pathology, Shanghai Jiaotong University Affiliated First People’s Hospital, 85 Wujin Road, Shanghai 200080, China; 5Department of General Surgery, Qianfoshan Hospital Affiliated to Shandong University, 16766 Jingshi Road, Jinan 250014, Shandong, China. *Equal contributors. Received August 21, 2014; Accepted September 15, 2014; Epub September 15, 2014; Published October 1, 2014 Abstract: EIF3e is a component of the eukaryotic translation initiation factor 3 (eIF-3) complexes, which is an es- sential factor for initiation of protein synthesis in mammalian cells. Translational control plays key roles in the complex mechanism of cancer development and progression. However, the clinical significance of eIF3e in colon cancer remains to be elucidated. We analyzed the eIF3e expression in a tissue microarray (TMA), which contained 173 colon cancer tissues paired with adjacent normal mucosa and lymph node metastasis. The expression of eIF3e was significantly elevated in colon cancer tissues in comparison with those in adjacent normal mucosa (P < 0.001) and lymph node metastasis (P < 0.001). -
Supplementary Tables Supplementary Table S1. a List of 3,039 Genes
Supplementary Tables Supplementary Table S1. A List of 3,039 Genes Correlated with HDAC2 Gene Expression from a HCC Large Cohort Study. AACS CSDA KLF15 PAFAH2 AASS CSE1L KLF9 PAH ABAT CSE1L KLHL2 PAK1IP1 ABCA2 CSE1L KLKB1 PAK4 ABCA5 CSNK1G1 KMO PAK6 ABCA5 CSNK2A1 KNG1 PAN3 ABCA6 CSNK2A1P KNTC1 PANK1 ABCA9 CSPP1 KPNA2 PAOX ABCA9 CTBP2 KPNA4 PAPOLA ABCB1 CTDSP2 KPNA5 PAPSS1 ABCB4 CTDSPL2 KPNB1 PAQR8 ABCC10 CTH KRAS PARD6A ABCC6 CTHRC1 KRT3 PARD6G ABCC6 CTNND2 L3MBTL3 PARP1 ABCC6 CTSB LACTB PARP10 ABCC9 CTSL2 LAMB1 PARP10 ABCF1 CTSO LAMC1 PARP3 ABCG5 CTXN1 LAMC2 PBLD ABCG8 CUX2 LAMP2 PBX1 ABHD15 CWC22 LAMP2 PBX2 ABHD2 CXXC6 LAPTM4B PCBD1 ABHD5 CYB5A LARP1B PCBP2 ABHD6 CYB5D2 LARP4 PCCA ABHD7 CYP11A1 LCAT PCCB ABL1 CYP26B1 LCN12 PCK1 ABTB2 CYP27A1 LCOR PCK2 ACAA1 CYP2A7 LDHD PCK2 ACAA2 CYP2C8 LDOC1 PCK2 ACACB CYP2C9 LDOC1L PCMT1 ACAD10 CYP2D6 LEAP-2 PCNA ACAD11 CYP2D7P1 LEMD3 PCNP ACADL CYP2J2 LEPR PCSK1N ACADM CYP2W1 LEPR PCSK6 ACADS CYP3A4 LEPR PCTP ACAT1 CYP3A43 LGALS2 PDCD2 ACBD3 CYP3A5 LGALS7 PDCD2 ACBD4 CYP4A11 LIMA1 PDCL3 ACBD5 CYP4A22 LIMS2 PDE5A ACCN3 CYP4F11 LINGO1 PDK2 ACMSD CYP4F12 LIPG PDK4 ACO1 CYP4F2 LMAN1 PDS5B ACOT1 CYP4F3 LMAN2 PEA15 ACOT12 CYP7B1 LMAN2L PEBP1 ACOT4 CYP8B1 LMCD1 PECR ACOT8 DAK LMNB2 PELI1 ACOX1 DAO LOC120376 PELP1 ACOX2 DARS LOC123876 PEMT ACPL2 DBF4 LOC127295 PEPD ACSL1 DBN1 LOC130773 PER1 ACSM2A DBNDD2 LOC136143 PEX11G ACSM2B DBNL LOC143543 PEX13 ACSM2B DBT LOC143941 PEX14 ACSM2B DCAF16 LOC146439 PFDN5 ACSM3 DCAF7 LOC147710 PFKFB1 ACSM3 DCDC2 LOC158345 PGF ACSM5 DCTN2 LOC201725 PGM1 ACSM5 DCXR LOC220433 -

Genes and Pathways Differentially Expressed in Bipolar Disorder And
(19) TZZ ¥Z _T (11) EP 2 453 024 B1 (12) EUROPEAN PATENT SPECIFICATION (45) Date of publication and mention (51) Int Cl.: of the grant of the patent: C12Q 1/68 (2006.01) G01N 33/53 (2006.01) 06.12.2017 Bulletin 2017/49 A61K 31/713 (2006.01) A61K 38/00 (2006.01) A61K 48/00 (2006.01) (21) Application number: 11175854.6 (22) Date of filing: 21.06.2005 (54) Genes and pathways differentially expressed in bipolar disorder and/or major depressive disorder Differenziell exprimierte Gene und Leitungsbahnen bei bipolarer Störung und/oder schwerer depressiver Erkrankung Gènes et voies exprimés différemment dans le trouble bipolaire et/ou le trouble dépressif majeur (84) Designated Contracting States: • Lopez, Juan F. AT BE BG CH CY CZ DE DK EE ES FI FR GB GR Ann Arbor, MI Michigan MI 48108 (US) HU IE IS IT LI LT LU MC NL PL PT RO SE SI SK TR • Lyons, David M. Palo Alto, CA California CA 94025 (US) (30) Priority: 21.06.2004 US 581998 P • Myers, Richard M. 22.10.2004 US 621252 P Stanford, CA California CA 94305 (US) 31.03.2005 US 667296 P • Schatzberg, Alan F. Los Altos, CA California CA 94024 (US) (43) Date of publication of application: • Stein, Richard 16.05.2012 Bulletin 2012/20 Irvine, CA 92620 (US) • Thompson, Robert C. (62) Document number(s) of the earlier application(s) in Ann Arbor, MI Michigan MI 48108 (US) accordance with Art. 76 EPC: • Tomita, Hiroaki 05762518.8 / 1 766 077 Sendai 989-3201 (JP) • Vawter, Marquis P. -
(12) Patent Application Publication (10) Pub. No.: US 2005/0287544A1 Bertucci Et Al
US 20050287544A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2005/0287544A1 Bertucci et al. (43) Pub. Date: Dec. 29, 2005 (54) GENE EXPRESSION PROFILING OF COLON Related U.S. Application Data CANCER WITH DNA ARRAYS (60) Provisional application No. 60/525,987, filed on Dec. 1, 2003. (76) Inventors: Francois Bertucci, Marseille (FR); Remi Houlgatte, Marseille (FR); Publication Classification Daniel Birnbaum, Marseille (FR); (51) Int. Cl. ................................................... C12O 1/68 Stephane Debono, Marseille (FR) (52) U.S. Cl. .................................................................. 435/6 (57) ABSTRACT Correspondence Address: IP GROUP OF DLA PIPER RUDNICK GRAY Differential gene expression associated with histopathologic CARY US LLP features of colorectal disease can be performed with nucleic 1650 MARKET ST acid arrayS. Such arrays can comprise a pool of polynucle SUTE 4900 otide Sequences from colon tissues, and the detection of the overexpression or underexpression of polynucleotide PHILADELPHIA, PA 19103 (US) Sequences (or Subsequences or complements thereof) from (21) Appl. No.: 11/000,688 this pool can provide information relating to the detection, diagnosis, Stage, classification, monitoring, prediction, pre (22) Filed: Dec. 1, 2004 vention or treatment of colorectal disease. Patent Application Publication Dec. 29, 2005 Sheet 1 of 5 US 2005/0287544A1 866 Meta - 899 EMeta - 785 Meta - 905 eta - D Group A 8031 feta - D 835 fetal - ) 8582 feta - ) 74421 Meta - D - 8208 letta - D 750S Eleta - A 8252. Roeta - A 9 Roeta - A 7943. Noneta - A 6992T ?eta - A 6952 Roeta - A Group B 692. Foeta - A 848 Roeta - A G94T Roleta - A 8646. Raeta - A 9118 Roeta - A 650 feta - A 694 NoMeta - A aeses FIG.