System Approaches Reveal the Molecular Networks Involved in Neural Stem Cell Differentiation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ARTICLE Doi:10.1038/Nature10523
ARTICLE doi:10.1038/nature10523 Spatio-temporal transcriptome of the human brain Hyo Jung Kang1*, Yuka Imamura Kawasawa1*, Feng Cheng1*, Ying Zhu1*, Xuming Xu1*, Mingfeng Li1*, Andre´ M. M. Sousa1,2, Mihovil Pletikos1,3, Kyle A. Meyer1, Goran Sedmak1,3, Tobias Guennel4, Yurae Shin1, Matthew B. Johnson1,Zˇeljka Krsnik1, Simone Mayer1,5, Sofia Fertuzinhos1, Sheila Umlauf6, Steven N. Lisgo7, Alexander Vortmeyer8, Daniel R. Weinberger9, Shrikant Mane6, Thomas M. Hyde9,10, Anita Huttner8, Mark Reimers4, Joel E. Kleinman9 & Nenad Sˇestan1 Brain development and function depend on the precise regulation of gene expression. However, our understanding of the complexity and dynamics of the transcriptome of the human brain is incomplete. Here we report the generation and analysis of exon-level transcriptome and associated genotyping data, representing males and females of different ethnicities, from multiple brain regions and neocortical areas of developing and adult post-mortem human brains. We found that 86 per cent of the genes analysed were expressed, and that 90 per cent of these were differentially regulated at the whole-transcript or exon level across brain regions and/or time. The majority of these spatio-temporal differences were detected before birth, with subsequent increases in the similarity among regional transcriptomes. The transcriptome is organized into distinct co-expression networks, and shows sex-biased gene expression and exon usage. We also profiled trajectories of genes associated with neurobiological categories and diseases, and identified associations between single nucleotide polymorphisms and gene expression. This study provides a comprehensive data set on the human brain transcriptome and insights into the transcriptional foundations of human neurodevelopment. -

Growth and Molecular Profile of Lung Cancer Cells Expressing Ectopic LKB1: Down-Regulation of the Phosphatidylinositol 3-Phosphate Kinase/PTEN Pathway1
[CANCER RESEARCH 63, 1382–1388, March 15, 2003] Growth and Molecular Profile of Lung Cancer Cells Expressing Ectopic LKB1: Down-Regulation of the Phosphatidylinositol 3-Phosphate Kinase/PTEN Pathway1 Ana I. Jimenez, Paloma Fernandez, Orlando Dominguez, Ana Dopazo, and Montserrat Sanchez-Cespedes2 Molecular Pathology Program [A. I. J., P. F., M. S-C.], Genomics Unit [O. D.], and Microarray Analysis Unit [A. D.], Spanish National Cancer Center, 28029 Madrid, Spain ABSTRACT the cell cycle in G1 (8, 9). However, the intrinsic mechanism by which LKB1 activity is regulated in cells and how it leads to the suppression Germ-line mutations in LKB1 gene cause the Peutz-Jeghers syndrome of cell growth is still unknown. It has been proposed that growth (PJS), a genetic disease with increased risk of malignancies. Recently, suppression by LKB1 is mediated through p21 in a p53-dependent LKB1-inactivating mutations have been identified in one-third of sporadic lung adenocarcinomas, indicating that LKB1 gene inactivation is critical in mechanism (7). In addition, it has been observed that LKB1 binds to tumors other than those of the PJS syndrome. However, the in vivo brahma-related gene 1 protein (BRG1) and this interaction is required substrates of LKB1 and its role in cancer development have not been for BRG1-induced growth arrest (10). Similar to what happens in the completely elucidated. Here we show that overexpression of wild-type PJS, Lkb1 heterozygous knockout mice show gastrointestinal hamar- LKB1 protein in A549 lung adenocarcinomas cells leads to cell-growth tomatous polyposis and frequent hepatocellular carcinomas (11, 12). suppression. To examine changes in gene expression profiles subsequent to Interestingly, the hamartomas, but not the malignant tumors, arising in exogenous wild-type LKB1 in A549 cells, we used cDNA microarrays. -

Supplementary Materials
Supplementary materials Supplementary Table S1: MGNC compound library Ingredien Molecule Caco- Mol ID MW AlogP OB (%) BBB DL FASA- HL t Name Name 2 shengdi MOL012254 campesterol 400.8 7.63 37.58 1.34 0.98 0.7 0.21 20.2 shengdi MOL000519 coniferin 314.4 3.16 31.11 0.42 -0.2 0.3 0.27 74.6 beta- shengdi MOL000359 414.8 8.08 36.91 1.32 0.99 0.8 0.23 20.2 sitosterol pachymic shengdi MOL000289 528.9 6.54 33.63 0.1 -0.6 0.8 0 9.27 acid Poricoic acid shengdi MOL000291 484.7 5.64 30.52 -0.08 -0.9 0.8 0 8.67 B Chrysanthem shengdi MOL004492 585 8.24 38.72 0.51 -1 0.6 0.3 17.5 axanthin 20- shengdi MOL011455 Hexadecano 418.6 1.91 32.7 -0.24 -0.4 0.7 0.29 104 ylingenol huanglian MOL001454 berberine 336.4 3.45 36.86 1.24 0.57 0.8 0.19 6.57 huanglian MOL013352 Obacunone 454.6 2.68 43.29 0.01 -0.4 0.8 0.31 -13 huanglian MOL002894 berberrubine 322.4 3.2 35.74 1.07 0.17 0.7 0.24 6.46 huanglian MOL002897 epiberberine 336.4 3.45 43.09 1.17 0.4 0.8 0.19 6.1 huanglian MOL002903 (R)-Canadine 339.4 3.4 55.37 1.04 0.57 0.8 0.2 6.41 huanglian MOL002904 Berlambine 351.4 2.49 36.68 0.97 0.17 0.8 0.28 7.33 Corchorosid huanglian MOL002907 404.6 1.34 105 -0.91 -1.3 0.8 0.29 6.68 e A_qt Magnogrand huanglian MOL000622 266.4 1.18 63.71 0.02 -0.2 0.2 0.3 3.17 iolide huanglian MOL000762 Palmidin A 510.5 4.52 35.36 -0.38 -1.5 0.7 0.39 33.2 huanglian MOL000785 palmatine 352.4 3.65 64.6 1.33 0.37 0.7 0.13 2.25 huanglian MOL000098 quercetin 302.3 1.5 46.43 0.05 -0.8 0.3 0.38 14.4 huanglian MOL001458 coptisine 320.3 3.25 30.67 1.21 0.32 0.9 0.26 9.33 huanglian MOL002668 Worenine -

Literature Mining Sustains and Enhances Knowledge Discovery from Omic Studies
LITERATURE MINING SUSTAINS AND ENHANCES KNOWLEDGE DISCOVERY FROM OMIC STUDIES by Rick Matthew Jordan B.S. Biology, University of Pittsburgh, 1996 M.S. Molecular Biology/Biotechnology, East Carolina University, 2001 M.S. Biomedical Informatics, University of Pittsburgh, 2005 Submitted to the Graduate Faculty of School of Medicine in partial fulfillment of the requirements for the degree of Doctor of Philosophy University of Pittsburgh 2016 UNIVERSITY OF PITTSBURGH SCHOOL OF MEDICINE This dissertation was presented by Rick Matthew Jordan It was defended on December 2, 2015 and approved by Shyam Visweswaran, M.D., Ph.D., Associate Professor Rebecca Jacobson, M.D., M.S., Professor Songjian Lu, Ph.D., Assistant Professor Dissertation Advisor: Vanathi Gopalakrishnan, Ph.D., Associate Professor ii Copyright © by Rick Matthew Jordan 2016 iii LITERATURE MINING SUSTAINS AND ENHANCES KNOWLEDGE DISCOVERY FROM OMIC STUDIES Rick Matthew Jordan, M.S. University of Pittsburgh, 2016 Genomic, proteomic and other experimentally generated data from studies of biological systems aiming to discover disease biomarkers are currently analyzed without sufficient supporting evidence from the literature due to complexities associated with automated processing. Extracting prior knowledge about markers associated with biological sample types and disease states from the literature is tedious, and little research has been performed to understand how to use this knowledge to inform the generation of classification models from ‘omic’ data. Using pathway analysis methods to better understand the underlying biology of complex diseases such as breast and lung cancers is state-of-the-art. However, the problem of how to combine literature- mining evidence with pathway analysis evidence is an open problem in biomedical informatics research. -

ITGB3BP (NM 014288) Human Recombinant Protein – TP300064
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TP300064 ITGB3BP (NM_014288) Human Recombinant Protein Product data: Product Type: Recombinant Proteins Description: Recombinant protein of human integrin beta 3 binding protein (beta3-endonexin) (ITGB3BP) Species: Human Expression Host: HEK293T Tag: C-Myc/DDK Predicted MW: 20 kDa Concentration: >50 ug/mL as determined by microplate BCA method Purity: > 80% as determined by SDS-PAGE and Coomassie blue staining Buffer: 25 mM Tris.HCl, pH 7.3, 100 mM glycine, 10% glycerol Preparation: Recombinant protein was captured through anti-DDK affinity column followed by conventional chromatography steps. Storage: Store at -80°C. Stability: Stable for 12 months from the date of receipt of the product under proper storage and handling conditions. Avoid repeated freeze-thaw cycles. RefSeq: NP_055103 Locus ID: 23421 UniProt ID: Q13352 RefSeq Size: 1019 Cytogenetics: 1p31.3 RefSeq ORF: 531 Synonyms: CENP-R; CENPR; HSU37139; NRIF3; TAP20 This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 2 ITGB3BP (NM_014288) Human Recombinant Protein – TP300064 Summary: This gene encodes a transcriptional coregulator that binds to and enhances the activity of members of the nuclear receptor families, thyroid hormone receptors and retinoid X receptors. This protein also acts as a corepressor of NF-kappaB-dependent signaling. This protein induces apoptosis in breast cancer cells through a caspase 2-mediated signaling pathway. -

A High-Throughput Approach to Uncover Novel Roles of APOBEC2, a Functional Orphan of the AID/APOBEC Family
Rockefeller University Digital Commons @ RU Student Theses and Dissertations 2018 A High-Throughput Approach to Uncover Novel Roles of APOBEC2, a Functional Orphan of the AID/APOBEC Family Linda Molla Follow this and additional works at: https://digitalcommons.rockefeller.edu/ student_theses_and_dissertations Part of the Life Sciences Commons A HIGH-THROUGHPUT APPROACH TO UNCOVER NOVEL ROLES OF APOBEC2, A FUNCTIONAL ORPHAN OF THE AID/APOBEC FAMILY A Thesis Presented to the Faculty of The Rockefeller University in Partial Fulfillment of the Requirements for the degree of Doctor of Philosophy by Linda Molla June 2018 © Copyright by Linda Molla 2018 A HIGH-THROUGHPUT APPROACH TO UNCOVER NOVEL ROLES OF APOBEC2, A FUNCTIONAL ORPHAN OF THE AID/APOBEC FAMILY Linda Molla, Ph.D. The Rockefeller University 2018 APOBEC2 is a member of the AID/APOBEC cytidine deaminase family of proteins. Unlike most of AID/APOBEC, however, APOBEC2’s function remains elusive. Previous research has implicated APOBEC2 in diverse organisms and cellular processes such as muscle biology (in Mus musculus), regeneration (in Danio rerio), and development (in Xenopus laevis). APOBEC2 has also been implicated in cancer. However the enzymatic activity, substrate or physiological target(s) of APOBEC2 are unknown. For this thesis, I have combined Next Generation Sequencing (NGS) techniques with state-of-the-art molecular biology to determine the physiological targets of APOBEC2. Using a cell culture muscle differentiation system, and RNA sequencing (RNA-Seq) by polyA capture, I demonstrated that unlike the AID/APOBEC family member APOBEC1, APOBEC2 is not an RNA editor. Using the same system combined with enhanced Reduced Representation Bisulfite Sequencing (eRRBS) analyses I showed that, unlike the AID/APOBEC family member AID, APOBEC2 does not act as a 5-methyl-C deaminase. -

The Genetic Program of Pancreatic Beta-Cell Replication in Vivo
Page 1 of 65 Diabetes The genetic program of pancreatic beta-cell replication in vivo Agnes Klochendler1, Inbal Caspi2, Noa Corem1, Maya Moran3, Oriel Friedlich1, Sharona Elgavish4, Yuval Nevo4, Aharon Helman1, Benjamin Glaser5, Amir Eden3, Shalev Itzkovitz2, Yuval Dor1,* 1Department of Developmental Biology and Cancer Research, The Institute for Medical Research Israel-Canada, The Hebrew University-Hadassah Medical School, Jerusalem 91120, Israel 2Department of Molecular Cell Biology, Weizmann Institute of Science, Rehovot, Israel. 3Department of Cell and Developmental Biology, The Silberman Institute of Life Sciences, The Hebrew University of Jerusalem, Jerusalem 91904, Israel 4Info-CORE, Bioinformatics Unit of the I-CORE Computation Center, The Hebrew University and Hadassah, The Institute for Medical Research Israel- Canada, The Hebrew University-Hadassah Medical School, Jerusalem 91120, Israel 5Endocrinology and Metabolism Service, Department of Internal Medicine, Hadassah-Hebrew University Medical Center, Jerusalem 91120, Israel *Correspondence: [email protected] Running title: The genetic program of pancreatic β-cell replication 1 Diabetes Publish Ahead of Print, published online March 18, 2016 Diabetes Page 2 of 65 Abstract The molecular program underlying infrequent replication of pancreatic beta- cells remains largely inaccessible. Using transgenic mice expressing GFP in cycling cells we sorted live, replicating beta-cells and determined their transcriptome. Replicating beta-cells upregulate hundreds of proliferation- related genes, along with many novel putative cell cycle components. Strikingly, genes involved in beta-cell functions, namely glucose sensing and insulin secretion were repressed. Further studies using single molecule RNA in situ hybridization revealed that in fact, replicating beta-cells double the amount of RNA for most genes, but this upregulation excludes genes involved in beta-cell function. -

Computational Simulations to Predict Creatine Kinase-Associated Factors: Protein-Protein Interaction Studies of Brain and Muscle Types of Creatine Kinases
SAGE-Hindawi Access to Research Enzyme Research Volume 2011, Article ID 328249, 12 pages doi:10.4061/2011/328249 Research Article Computational Simulations to Predict Creatine Kinase-Associated Factors: Protein-Protein Interaction Studies of Brain and Muscle Types of Creatine Kinases Wei-Jiang Hu,1 Sheng-Mei Zhou,2 Joshua SungWoo Yang,3, 4 and Fan-Guo Meng1 1 Zhejiang Provincial Key Laboratory of Applied Enzymology, Yangtze Delta Region Institute of Tsinghua University, Jiaxing 314006, China 2 College of Biology and Chemical Engineering, Jiaxing University, Jiaxing 314001, China 3 Korean Bioinformation Center (KOBIC), Korea Research Institute of Bioscience & Biotechnology (KRIBB), Daejeon 305-806, Republic of Korea 4 Department of Bioinformatics, University of Sciences & Technology, Daejeon 205-305, Republic of Korea Correspondence should be addressed to Fan-Guo Meng, [email protected] Received 17 May 2011; Accepted 26 May 2011 Academic Editor: Jun-Mo Yang Copyright © 2011 Wei-Jiang Hu et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Creatine kinase (CK; EC 2.7.3.2) is related to several skin diseases such as psoriasis and dermatomyositis. CK is important in skin energy homeostasis because it catalyzes the reversible transfer of a phosphoryl group from MgATP to creatine. In this study, we predicted CK binding proteins via the use of bioinformatic tools such as protein-protein interaction (PPI) mappings and suggest the putative hub proteins for CK interactions. We obtained 123 proteins for brain type CK and 85 proteins for muscle type CK in the interaction networks. -
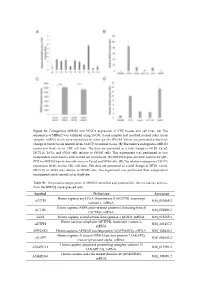
Figure S1. Endogenous MIR45
Figure S1. Endogenous MIR452 and VEGFA expression in CRC tissues and cell lines. (A) The expression of MIR452 was validated using 10 CRC tissue samples and matched normal colon tissue samples. miRNA levels were normalized to colon-specific RNU48. Values are presented as the fold- change in tumor tissue relative levels (ΔΔCT) to normal tissue. (B) The relative endogenous MIR452 expression levels in six CRC cell lines. The data are presented as a fold change in HT29, Caco2, HCT116, LoVo, and SW48 cells relative to SW480 cells. This experiment was performed as two independent experiments, each carried out in triplicate. (C) MIR452 expression level analysis by qRT- PCR for MIR452 transfection efficiency in Caco2 and SW48 cells. (D) The relative endogenous VEGFA expression levels in five CRC cell lines. The data are presented as a fold change in HT29, Caco2, HCT116, or SW48 cells relative to SW480 cells. This experiment was performed three independent experiments, each carried out in duplicate. Table S1. The putative target genes of MIR452 identified and predicted by the microarray analysis from the MIR452 overexpressed cells. Symbol Definition Accession Homo sapiens acyl-CoA thioesterase 8 (ACOT8), transcript ACOT8 NM_005469.2 variant 1, mRNA. Homo sapiens ARP6 actin-related protein 6 homolog (yeast) ACTR6 NM_022496.3 (ACTR6), mRNA. ADI1 Homo sapiens acireductone dioxygenase 1 (ADI1), mRNA. NM_018269.1 Homo sapiens aftiphilin (AFTPH), transcript variant 1, AFTPH NM_203437.2 mRNA. AHNAK2 Homo sapiens AHNAK nucleoprotein 2 (AHNAK2), mRNA. NM_138420.2 Homo sapiens A kinase (PRKA) anchor protein 7 (AKAP7), AKAP7 NM_004842.2 transcript variant alpha, mRNA. Homo sapiens anaphase promoting complex subunit 13 ANAPC13 NM_015391.2 (ANAPC13), mRNA. -

Mouse Itgb3bp Knockout Project (CRISPR/Cas9)
https://www.alphaknockout.com Mouse Itgb3bp Knockout Project (CRISPR/Cas9) Objective: To create a Itgb3bp knockout Mouse model (C57BL/6J) by CRISPR/Cas-mediated genome engineering. Strategy summary: The Itgb3bp gene (NCBI Reference Sequence: NM_026348 ; Ensembl: ENSMUSG00000028549 ) is located on Mouse chromosome 4. 9 exons are identified, with the ATG start codon in exon 1 and the TGA stop codon in exon 8 (Transcript: ENSMUST00000146258). Exon 3~4 will be selected as target site. Cas9 and gRNA will be co-injected into fertilized eggs for KO Mouse production. The pups will be genotyped by PCR followed by sequencing analysis. Note: Exon 3 starts from about 9.28% of the coding region. Exon 3~4 covers 38.45% of the coding region. The size of effective KO region: ~3537 bp. The KO region does not have any other known gene. Page 1 of 9 https://www.alphaknockout.com Overview of the Targeting Strategy Wildtype allele 5' gRNA region gRNA region 3' 1 3 4 9 Legends Exon of mouse Itgb3bp Knockout region Page 2 of 9 https://www.alphaknockout.com Overview of the Dot Plot (up) Window size: 15 bp Forward Reverse Complement Sequence 12 Note: The 2000 bp section upstream of Exon 3 is aligned with itself to determine if there are tandem repeats. No significant tandem repeat is found in the dot plot matrix. So this region is suitable for PCR screening or sequencing analysis. Overview of the Dot Plot (down) Window size: 15 bp Forward Reverse Complement Sequence 12 Note: The 2000 bp section downstream of Exon 4 is aligned with itself to determine if there are tandem repeats. -

A Microrna Program Regulates the Balance Between Cardiomyocyte Hyperplasia and Hypertrophy and Stimulates Cardiac Regeneration
ARTICLE https://doi.org/10.1038/s41467-021-25211-4 OPEN A microRNA program regulates the balance between cardiomyocyte hyperplasia and hypertrophy and stimulates cardiac regeneration Andrea Raso1,12, Ellen Dirkx1,12, Vasco Sampaio-Pinto1,2, Hamid el Azzouzi1,3, Ryan J. Cubero 4,5, Daniel W. Sorensen6, Lara Ottaviani1, Servé Olieslagers1, Manon M. Huibers 7, Roel de Weger7, Sailay Siddiqi8, Silvia Moimas 9, Consuelo Torrini9, Lorena Zentillin9, Luca Braga9, Diana S. Nascimento 2, Paula A. da Costa Martins 1,10, Jop H. van Berlo 6, Serena Zacchigna 8, Mauro Giacca 9,11 & ✉ Leon J. De Windt 1 1234567890():,; Myocardial regeneration is restricted to early postnatal life, when mammalian cardiomyo- cytes still retain the ability to proliferate. The molecular cues that induce cell cycle arrest of neonatal cardiomyocytes towards terminally differentiated adult heart muscle cells remain obscure. Here we report that the miR-106b~25 cluster is higher expressed in the early postnatal myocardium and decreases in expression towards adulthood, especially under conditions of overload, and orchestrates the transition of cardiomyocyte hyperplasia towards cell cycle arrest and hypertrophy by virtue of its targetome. In line, gene delivery of miR- 106b~25 to the mouse heart provokes cardiomyocyte proliferation by targeting a network of negative cell cycle regulators including E2f5, Cdkn1c, Ccne1 and Wee1. Conversely, gene- targeted miR-106b~25 null mice display spontaneous hypertrophic remodeling and exag- gerated remodeling to overload by derepression of the prohypertrophic transcription factors Hand2 and Mef2d. Taking advantage of the regulatory function of miR-106b~25 on cardio- myocyte hyperplasia and hypertrophy, viral gene delivery of miR-106b~25 provokes nearly complete regeneration of the adult myocardium after ischemic injury. -

Mouse Endogenous Retroviruses Can Trigger Premature Transcriptional Termination at a Distance
Downloaded from genome.cshlp.org on September 28, 2021 - Published by Cold Spring Harbor Laboratory Press Research Mouse endogenous retroviruses can trigger premature transcriptional termination at a distance Jingfeng Li,1 Keiko Akagi,1 Yongjun Hu,2 Anna L. Trivett,3 Christopher J.W. Hlynialuk,1 Deborah A. Swing,4 Natalia Volfovsky,5 Tamara C. Morgan,6 Yelena Golubeva,6 Robert M. Stephens,5 David E. Smith,2 and David E. Symer1,7,8 1Human Cancer Genetics Program and Department of Molecular Virology, Immunology and Medical Genetics, The Ohio State University Comprehensive Cancer Center, Columbus, Ohio 43210, USA; 2Department of Pharmaceutical Sciences, University of Michigan, Ann Arbor, Michigan 48109, USA; 3Laboratory of Molecular Immunoregulation and 4Mouse Cancer Genetics Program, National Cancer Institute, Frederick, Maryland 21702, USA; 5Advanced Biomedical Computing Center, Information Systems Program and 6Histotechnology Laboratory, SAIC-Frederick, Inc., National Cancer Institute, Frederick, Maryland 21702, USA; 7Department of Internal Medicine and Department of Biomedical Informatics, The Ohio State University Comprehensive Cancer Center, Columbus, Ohio 43210, USA Endogenous retrotransposons have caused extensive genomic variation within mammalian species, but the functional implications of such mobilization are mostly unknown. We mapped thousands of endogenous retrovirus (ERV) germline integrants in highly divergent, previously unsequenced mouse lineages, facilitating a comparison of gene expression in the presence or absence of