Molecular Phylogeny and Taxonomy of Lepidoptera with Special Reference to Influence
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Evolution of Insect Color Vision: from Spectral Sensitivity to Visual Ecology
EN66CH23_vanderKooi ARjats.cls September 16, 2020 15:11 Annual Review of Entomology Evolution of Insect Color Vision: From Spectral Sensitivity to Visual Ecology Casper J. van der Kooi,1 Doekele G. Stavenga,1 Kentaro Arikawa,2 Gregor Belušic,ˇ 3 and Almut Kelber4 1Faculty of Science and Engineering, University of Groningen, 9700 Groningen, The Netherlands; email: [email protected] 2Department of Evolutionary Studies of Biosystems, SOKENDAI Graduate University for Advanced Studies, Kanagawa 240-0193, Japan 3Department of Biology, Biotechnical Faculty, University of Ljubljana, 1000 Ljubljana, Slovenia; email: [email protected] 4Lund Vision Group, Department of Biology, University of Lund, 22362 Lund, Sweden; email: [email protected] Annu. Rev. Entomol. 2021. 66:23.1–23.28 Keywords The Annual Review of Entomology is online at photoreceptor, compound eye, pigment, visual pigment, behavior, opsin, ento.annualreviews.org anatomy https://doi.org/10.1146/annurev-ento-061720- 071644 Abstract Annu. Rev. Entomol. 2021.66. Downloaded from www.annualreviews.org Copyright © 2021 by Annual Reviews. Color vision is widespread among insects but varies among species, depend- All rights reserved ing on the spectral sensitivities and interplay of the participating photore- Access provided by University of New South Wales on 09/26/20. For personal use only. ceptors. The spectral sensitivity of a photoreceptor is principally determined by the absorption spectrum of the expressed visual pigment, but it can be modified by various optical and electrophysiological factors. For example, screening and filtering pigments, rhabdom waveguide properties, retinal structure, and neural processing all influence the perceived color signal. -

Region of Nymphalidae and Libytheidae (Lepidoptera) from Japan
MORPHOLOGICAL STUDIES ON THE OCCIPITAL REGION OF NYMPHALIDAE AND LIBYTHEIDAE (LEPIDOPTERA) FROM JAPAN Takashi TSUBUKI and Nagao KOYAMA * .h2monji junior High School, 10-33, Kita6tsz{ha 1, Toshima-feu, Toleyo (170), 1opan ** Biological Laboratory, Shinshi2 Udeiversity, Cllada (386), 14pan CONTENTS I. Introduction・--・ny・・・・・・・・・・・・・・・・・-・-・・・・・・-・---・--t・-・---・ny・・・・-・-・・・-・-・・-・・・1 II. Materials and methods・-・・・・・・・・・t・・・・・・t・・・・・・・・・・・・・・`・・・・・・・・・・-・・・-・・・・・・-d・・・・・・・・・・・・2 III. General morphology of the occipital region ・・・・・・}・・・・・・・・・・・・・・・・・・・・・・・・・・・・・4 IV. Occipital structure of each species in Nymphaliclae and Libytheidae ・-ib-・・・・-・・・-・ny・・・・・--・-・・・・・・・-・・-・----・b-・・・----・-・・・-・・-・--・・-6 V. Phylogenical grouping of Nymphalidae based on the occipital structure・t・・・・・・・--・--・・-・-・・・・・・・・・・-・・・・-ny-・・・ny-・--・-----・H"""""".","".."."lg VI. Summary ・・・・-・-・-・・・・・・-・・----・・・-・・・・-・-・-・-----・・-------・-・----・23 VII. Literatures cited ・・・--・・・・・・・・・`・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・・--・-・・-・・・・・・tt・・・・・・23 I. INTRODUCTION A considerable number of studies have been carried out on the occipital morphology of moths since YAGI and KoYAMA's report of 1963 (KoYAMA and MlyATA, 1969, 1970, 1975;MIYATA, 1971, 1972, 1973, 1974, 1975;MIyATA and KOYAMA, 1971, 1972, 1976). In the butterfiies, however, few papers were available concerning the occipital region, on which EHRucH (1958, a b), YAGI and KoyAMA (1963) and KoyAMA and OGAwA (1972) briefly described. Then, tried to study the occipital region of butterflies, the authors observed it preliminarily (TsuBuKI et aL 1975). The present paper deals with the occipital structure of Nymphalidae and Libytheidae from Japan with reference to its bearing on systematics. Before going further, the authors wish to express their gratitude to Prof. Dr. H. SAwADA and Prof. Dr. N. GOKAN, Tokyo University of Agriculture, for their advice and assistance. Thanks are also due to Mr. N. K6DA who gave valuable materials for this work, and to Mr. Y. YAGucm and the members of JEOL Ltd. -
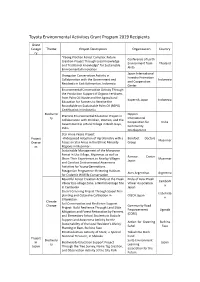
List of Previous Grant Projects
Toyota Environmental Activities Grant Program 2019 Recipients Grant Catego Theme Project Description Organization Country ry "Kaeng Krachan Forest Complex: Future Conference of Earth Creation Project Through Local Knowledge Environment from Thailand and Traditional Knowledge" for Sustainable Akita Environmental Innovation Japan International Orangutan Conservation Activity in Forestry Promotion Collaboration with the Government and Indonesia and Cooperation Residents in East Kalimantan, Indonesia Center Environmental Conservation Activity Through the Production Support of Organic Fertilizers from Palm Oil Waste and the Agricultural Kopernik Japan Indonesia Education for Farmers to Receive the Roundtable on Sustainable Palm Oil (RSPO) Certification in Indonesia Biodiversi Nippon Practical Environmental Education Project in ty International Collaboration with Children, Women, and the Cooperation for India Government in a Rural Village in Bodh Gaya, Community India Development Star Anise Peace Project Project -Widespread Adoption of Agroforestry with a Barefoot Doctors Myanmar Overse Focus on Star Anise in the Ethnic Minority Group as Regions in Myanmar- Sustainable Management of the Mangrove Forest in Uto Village, Myanmar, as well as Ramsar Center Share Their Experiences to Nearby Villages Myanmar Japan and Conduct Environmental Awareness Activities for Young Generations Patagonian Programme: Restoring Habitats Aves Argentinas Argentina for Endemic Wildlife Conservation Beautiful Forest Creation Activity at the Preah Pride of Asia: Preah -

Title Flowering Phenology and Anthophilous Insect Community at a Threatened Natural Lowland Marsh at Nakaikemi in Tsuruga, Japan
Flowering phenology and anthophilous insect community at a Title threatened natural lowland marsh at Nakaikemi in Tsuruga, Japan Author(s) KATO, Makoto; MIURA, Reiichi Contributions from the Biological Laboratory, Kyoto Citation University (1996), 29(1): 1 Issue Date 1996-03-31 URL http://hdl.handle.net/2433/156114 Right Type Departmental Bulletin Paper Textversion publisher Kyoto University Contr. biol. Lab. Kyoto Univ., Vol. 29, pp. 1-48, Pl. 1 Issued 31 March 1996 Flowering phenology and anthophilous insect community at a threatened natural lowland marsh at Nakaikemi in Tsuruga, Japan Makoto KATo and Reiichi MiuRA ABSTRACT Nakaikemi marsh, located in Fukui Prefecture, is one of only a few natural lowland marshlands left in westem Japan, and harbors many endangered marsh plants and animals. Flowering phenology and anthophilous insect communities on 64 plant species of 35 families were studied in the marsh in 1994-95. A total of 936 individuals of 215 species in eight orders of Insecta were collected on flowers from mid April to mid October, The anthophilous insect community was characterized by dominance of Diptera (58 9e of individuals) and relative paucity of Hymenoptera (26 9o), Hemiptera (6 9e), Lepidoptera (5 9e), and Coleoptera (5 9o), Syrphidae was the most abundant family and probably the most important pollination agents. Bee community was characterized by dominance of an aboveground nesting bee genus, Hylaeus (Colletidae), the most abundant species of which was a minute, rare little-recorded species. Cluster analysis on fiower-visiting insect spectra grouped 64 plant species into seven clusters, which were respectively characterized by dominance of small or large bees (18 spp.), syrphid fiies (13 spp.), Calyptrate and other flies (11 spp.), wasps and middle-sized bees (8 spp.), Lepidoptera (2 spp.), Coleoptera (1 sp.) and a mixture of these various insects (11 spp.). -

Hesperüdae of Vietnam, 151 New Records of Hesperiidae from Southern Vietnam (Lepidoptera, Hesperüdae) by A
ZOBODAT - www.zobodat.at Zoologisch-Botanische Datenbank/Zoological-Botanical Database Digitale Literatur/Digital Literature Zeitschrift/Journal: Atalanta Jahr/Year: 2003 Band/Volume: 34 Autor(en)/Author(s): Devyatkin Alexey L., Monastyrskii Alexander L. Artikel/Article: Hesperiidae of Vietnam, 15 New records of Hesperiidae from southern Vietnam (Lepidoptera, Hesperiidae) 119-133 ©Ges. zur Förderung d. Erforschung von Insektenwanderungen e.V. München, download unter www.zobodat.at Atalanta (August 2003) 34(1/2): 119-133, colour plate Xc, Würzburg, ISSN 0171-0079 Hesperüdae of Vietnam, 151 New records of Hesperiidae from southern Vietnam (Lepidoptera, Hesperüdae) by A. L.D evyatkin & A. L Monastyrskii received 5.V.2003 Summary: A total of 67 species is added to the list of Hesperiidae of southern Vietnam, 15 of them being new for the country as a whole. A new subspecies, Pyroneura callineura natalia subspec. nov. is described and illustrated. Taxonomic notes on certain species are presented. Since the previous publication summarizing the knowledge of the Hesperiidae in the southern part of Vietnam (Devyatkin & M onastyrskii , 2000), several further localities have been visited by research expeditions and individual collectors. The annotated list below is based predominantly on the material collected in the Cat Tien Na ture Reserve in 2000 (no year is given for the label data in the list), which was most profoundly studied and proved to be very rich and diverse in terms of the butterfly fauna, and contains new records for the south of the country along with some taxonomic corrections made in view of the new data. Although some of the areas concerned in this paper may be geographically attributed to the southern part of Central Vietnam (or Annam), they were not regarded in our previous publica tions dedicated to the northern and central areas of the country (Devyatkin & M onastyrskii , 1999, 2002), the new data thus being supplementary to those published before on the south ern part of Vietnam (Devyatkin & M onastyrskii , 2000). -

The Taxonomy of the Japanese Oak Red Scale Insect
Zootaxa 3630 (2): 291–307 ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ Article ZOOTAXA Copyright © 2013 Magnolia Press ISSN 1175-5334 (online edition) http://dx.doi.org/10.11646/zootaxa.3630.2.5 http://zoobank.org/urn:lsid:zoobank.org:pub:373402BA-36A9-4323-AE9D-A84D89C47231 The taxonomy of the Japanese oak red scale insect, Kuwania quercus (Kuwana) (Hemiptera: Coccoidea: Kuwaniidae), with a generic diagnosis, a key to species and description of a new species from California SAN’AN WU1, NAN NAN1, PENNY GULLAN2 & JUN DENG1 1The Key Laboratory for Silviculture and Conservation of Ministry of Education, Beijing Forestry University, Beijing, P. R. China 100083. E-mail: [email protected] 2Division of Evolution, Ecology and Genetics, Research School of Biology, The Australian National University, Canberra, A.C.T. 0200, Australia. E-mail: [email protected] Abstract The oak red scale insect, Kuwania quercus (Kuwana), was described from specimens collected from the bark of oak trees (Quercus species) in Japan. More recently, the species has been identified from California and China, but Californian specimens differ morphologically from Japanese material and are considered here to be a new species based on both morphological and molecular data. In this paper, an illustrated redescription of K. quercus is provided based on type specimens consisting of adult females, first-instar nymphs and intermediate-stage females, and a lectotype is designated for Sasakia quercus Kuwana. The new Californian species, Kuwania raygilli Wu & Gullan, is described and illustrated based on the adult female, first-instar nymph and intermediate-stage female. A new generic diagnosis for Kuwania Cockerell based on adult females and first-instar nymphs, and a key to species based on adult females are included. -

Mai Po Nature Reserve Management Plan: 2019-2024
Mai Po Nature Reserve Management Plan: 2019-2024 ©Anthony Sun June 2021 (Mid-term version) Prepared by WWF-Hong Kong Mai Po Nature Reserve Management Plan: 2019-2024 Page | 1 Table of Contents EXECUTIVE SUMMARY ................................................................................................................................................... 2 1. INTRODUCTION ..................................................................................................................................................... 7 1.1 Regional and Global Context ........................................................................................................................ 8 1.2 Local Biodiversity and Wise Use ................................................................................................................... 9 1.3 Geology and Geological History ................................................................................................................. 10 1.4 Hydrology ................................................................................................................................................... 10 1.5 Climate ....................................................................................................................................................... 10 1.6 Climate Change Impacts ............................................................................................................................. 11 1.7 Biodiversity ................................................................................................................................................ -

Molecular Phylogeny of the Genus Satarupa Moore
國立臺灣師範大學生命科學系碩士論文 颯弄蝶屬之分子親緣與系統研究 Molecular phylogeny and systematics of the genus Satarupa Moore, 1865 (Lepidoptera: Hesperiidae: Pyrginae) 研究生: 莊懷淳 Huai-chun Chuang 指導教授: 徐堉峰 博士 Yu-Feng Hsu 千葉秀幸 博士 Hideyuki Chiba 中華民國 102 年 7 月 致謝 首先要感謝我的父母,經過多番的波折與衝突答應讓我長久以來的心 願能夠一邊實驗一邊做田野調查。並且非常的感謝我的論文指導教 授,徐堉峰教授,給予我如此自由的環境讓我實驗。並且也要感謝實 驗室的各位夥伴,讓整個實驗室充滿了活潑的環境讓原本枯燥的研究 過程增添豐富的色彩,猶如雨天過後的七彩彩虹。感謝實驗室的學長 姐們帶領我野外調查並且獲得台灣難以獲得的樣本。並且也要感謝提 供我國外樣本的千葉秀幸博士,讓我獲得許多困難獲取的樣本,以及 提供我很多論文寫作的相關意見。當然一定要感謝提供如此高規格的 實驗環境的李壽先老師,並且在很多思考上給予很大的啟發,並且教 導我在很多表達上的細節與邏輯。感謝林思民老師在口試與報告的時 候給予我許多中肯的意見。也要感謝共同陪我奮鬥的同學們,一起在 實驗室熬夜趕 ppt 等口頭報告。最後最後一定要感謝這兩年來一起共 同奮鬥的顏嘉瑩同學,在這兩年一起度過了非常多的挫折與試煉,如 果沒有他我這兩年的色彩會黯淡許多。要感謝的人太多,寥寥字詞無 法述說滿滿的感謝,就如陳之藩所言:「要感謝的人太多,不如謝天 吧!」 Contents 中文摘要…………………………...……………………..……1 Abstract…………………………………………………..……2 Introduction………………………………….………….….…4 Materials and methods…………………………………….…8 Results……………………………………………………..….12 Discussion……………………………………………….……14 References...………………………………………………..…18 Tables………………………………………………….……...25 Figures………………………………………………….…….29 Appendix……………………………….……………………37 中文摘要 親緣關係為生物研究的基礎,弄蝶親緣關係已有高階親緣關係發表, 在屬級的親緣關係尚有不足之處。颯弄蝶屬屬特徵為前翅 2A 至 R3 脈之間有透明的斑紋與後翅具有大片的白塊。因颯弄蝶種之間的形態 太過於相似,許多發表尚有辨識錯誤的情形。Evans 在 1949 整理為 7 種,Okano (1987) 和 Chiba (1989) 則認為颯弄蝶屬為 valentini, zulla, gopala, nymphalis, splendens, monbeigi 和 formosibia。以翅形與幼蟲型 態 S. formosibia 與 S. monbeigi 應屬於同一類群,但是前者外生殖器卻 並不典型。Shirôzu 將 S. majasra 處理為 gopala 的亞種,但 Tsukiyama 處理為 nymphalis 的亞種。颯弄蝶屬近緣屬皆為熱帶分布,颯弄蝶卻 分布自爪哇至溫帶的西伯利亞。建構颯弄蝶屬親緣關係可以確立 formosibia 及 majasra 的分類地位並且了解颯弄蝶屬的溫帶分布是由 熱帶分布至溫帶或原本分布在溫帶。本研究使用 COI, COII 與 Ef1α -

Out of the Orient: Post-Tethyan Transoceanic and Trans-Arabian Routes
Systematic Entomology Page 2 of 55 1 1 Out of the Orient: Post-Tethyan transoceanic and trans-Arabian routes 2 fostered the spread of Baorini skippers in the Afrotropics 3 4 Running title: Historical biogeography of Baorini skippers 5 6 Authors: Emmanuel F.A. Toussaint1,2*, Roger Vila3, Masaya Yago4, Hideyuki Chiba5, Andrew 7 D. Warren2, Kwaku Aduse-Poku6,7, Caroline Storer2, Kelly M. Dexter2, Kiyoshi Maruyama8, 8 David J. Lohman6,9,10, Akito Y. Kawahara2 9 10 Affiliations: 11 1 Natural History Museum of Geneva, CP 6434, CH 1211 Geneva 6, Switzerland 12 2 Florida Museum of Natural History, University of Florida, Gainesville, Florida, 32611, U.S.A. 13 3 Institut de Biologia Evolutiva (CSIC-UPF), Passeig Marítim de la Barceloneta, 37, 08003 14 Barcelona, Spain 15 4 The University Museum, The University of Tokyo, Hongo, Bunkyo-ku, Tokyo 113-0033, Japan 16 5 B. P. Bishop Museum, 1525 Bernice Street, Honolulu, Hawaii, 96817-0916 U.S.A. 17 6 Biology Department, City College of New York, City University of New York, 160 Convent 18 Avenue, NY 10031, U.S.A. 19 7 Biology Department, University of Richmond, Richmond, Virginia, 23173, USA 20 8 9-7-106 Minami-Ôsawa 5 chome, Hachiôji-shi, Tokyo 192-0364, Japan 21 9 Ph.D. Program in Biology, Graduate Center, City University of New York, 365 Fifth Ave., New 22 York, NY 10016, U.S.A. 23 10 Entomology Section, National Museum of the Philippines, Manila 1000, Philippines 24 25 *To whom correspondence should be addressed: E-mail: [email protected] Page 3 of 55 Systematic Entomology 2 26 27 ABSTRACT 28 The origin of taxa presenting a disjunct distribution between Africa and Asia has puzzled 29 biogeographers for centuries. -

Lepidoptera: Noctuoidea: Erebidae) and Its Phylogenetic Implications
EUROPEAN JOURNAL OF ENTOMOLOGYENTOMOLOGY ISSN (online): 1802-8829 Eur. J. Entomol. 113: 558–570, 2016 http://www.eje.cz doi: 10.14411/eje.2016.076 ORIGINAL ARTICLE Characterization of the complete mitochondrial genome of Spilarctia robusta (Lepidoptera: Noctuoidea: Erebidae) and its phylogenetic implications YU SUN, SEN TIAN, CEN QIAN, YU-XUAN SUN, MUHAMMAD N. ABBAS, SAIMA KAUSAR, LEI WANG, GUOQING WEI, BAO-JIAN ZHU * and CHAO-LIANG LIU * College of Life Sciences, Anhui Agricultural University, 130 Changjiang West Road, Hefei, 230036, China; e-mails: [email protected] (Y. Sun), [email protected] (S. Tian), [email protected] (C. Qian), [email protected] (Y.-X. Sun), [email protected] (M.-N. Abbas), [email protected] (S. Kausar), [email protected] (L. Wang), [email protected] (G.-Q. Wei), [email protected] (B.-J. Zhu), [email protected] (C.-L. Liu) Key words. Lepidoptera, Noctuoidea, Erebidae, Spilarctia robusta, phylogenetic analyses, mitogenome, evolution, gene rearrangement Abstract. The complete mitochondrial genome (mitogenome) of Spilarctia robusta (Lepidoptera: Noctuoidea: Erebidae) was se- quenced and analyzed. The circular mitogenome is made up of 15,447 base pairs (bp). It contains a set of 37 genes, with the gene complement and order similar to that of other lepidopterans. The 12 protein coding genes (PCGs) have a typical mitochondrial start codon (ATN codons), whereas cytochrome c oxidase subunit 1 (cox1) gene utilizes unusually the CAG codon as documented for other lepidopteran mitogenomes. Four of the 13 PCGs have incomplete termination codons, the cox1, nad4 and nad6 with a single T, but cox2 has TA. It comprises six major intergenic spacers, with the exception of the A+T-rich region, spanning at least 10 bp in the mitogenome. -

The Taxonomy of the Japanese Oak Red Scale Insect
Zootaxa 3630 (2): 291–307 ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ Article ZOOTAXA Copyright © 2013 Magnolia Press ISSN 1175-5334 (online edition) http://dx.doi.org/10.11646/zootaxa.3630.2.5 http://zoobank.org/urn:lsid:zoobank.org:pub:373402BA-36A9-4323-AE9D-A84D89C47231 The taxonomy of the Japanese oak red scale insect, Kuwania quercus (Kuwana) (Hemiptera: Coccoidea: Kuwaniidae), with a generic diagnosis, a key to species and description of a new species from California SAN’AN WU1, NAN NAN1, PENNY GULLAN2 & JUN DENG1 1The Key Laboratory for Silviculture and Conservation of Ministry of Education, Beijing Forestry University, Beijing, P. R. China 100083. E-mail: [email protected] 2Division of Evolution, Ecology and Genetics, Research School of Biology, The Australian National University, Canberra, A.C.T. 0200, Australia. E-mail: [email protected] Abstract The oak red scale insect, Kuwania quercus (Kuwana), was described from specimens collected from the bark of oak trees (Quercus species) in Japan. More recently, the species has been identified from California and China, but Californian specimens differ morphologically from Japanese material and are considered here to be a new species based on both morphological and molecular data. In this paper, an illustrated redescription of K. quercus is provided based on type specimens consisting of adult females, first-instar nymphs and intermediate-stage females, and a lectotype is designated for Sasakia quercus Kuwana. The new Californian species, Kuwania raygilli Wu & Gullan, is described and illustrated based on the adult female, first-instar nymph and intermediate-stage female. A new generic diagnosis for Kuwania Cockerell based on adult females and first-instar nymphs, and a key to species based on adult females are included. -

The Mitogenome of a Malagasy Butterfly Malaza Fastuosus (Mabille
Title The mitogenome of a Malagasy butterfly Malaza fastuosus (Mabille, 1884) recovered from the holotype collected over 140 years ago adds support for a new subfamily of Hesperiidae (Lepidoptera) Authors Zhang, J; Lees, David; Shen, J; Cong, Q; Huertas, B; Martin, G; Grishin, NV 195 ARTICLE The mitogenome of a Malagasy butterfly Malaza fastuosus (Mabille, 1884) recovered from the holotype collected over 140 years ago adds support for a new subfamily of Hesperiidae (Lepidoptera) Jing Zhang, David C. Lees, Jinhui Shen, Qian Cong, Blanca Huertas, Geoff Martin, and Nick V. Grishin Abstract: Malaza fastuosus is a lavishly patterned skipper butterfly from a genus that has three described species, all endemic to the mainland of Madagascar. To our knowledge, M. fastuosus has not been collected for nearly 50 years. To evaluate the power of our techniques to recover DNA, we used a single foreleg of an at least 140-year-old holotype specimen from the collection of the Natural History Museum London with no destruction of external morphology to extract DNA and assemble a complete mitogenome from next generation sequencing reads. The resulting 15 540 bp mitogenome contains 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes, and an A+T rich region, similarly to other Lepidoptera mitogenomes. Here we provide the first mitogenome also for Trapezitinae (Rachelia extrusus). Phylogenetic analysis of available skipper mitogenomes places Malaza outside of Trapezitinae and Barcinae + Hesperiinae, with a possible sister relationship to Heteropterinae. Of these, at least Heteropterinae, Trape- zitinae, and almost all Hesperiinae have monocot-feeding caterpillars. Malaza appears to be an evolutionarily highly distinct ancient lineage, morphologically with several unusual hesperiid features.