Microbiome Profile of the Amniotic Fluid As a Predictive Biomarker Of
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

BD™ Gardnerella Selective Agar with 5% Human Blood
INSTRUCTIONS FOR USE – READY-TO-USE PLATED MEDIA PA-254094.06 Rev.: July 2014 BD Gardnerella Selective Agar with 5% Human Blood INTENDED USE BD Gardnerella Selective Agar with 5% Human Blood is a partially selective and differential medium for the isolation of Gardnerella vaginalis from clinical specimens. PRINCIPLES AND EXPLANATION OF THE PROCEDURE Microbiological method. Gardnerella vaginalis is considered to be one of the organisms causing vaginitis.1-4 Although the organism may be present in a high percentage of normal women in the vaginal flora, its importance as a cause of non-specific vaginitis (also called bacterial vaginosis) has never been questioned. In symptomatic women, G. vaginalis frequently is associated with anaerobes such as Prevotella bivia, P. disiens, Mobiluncus, Peptostreptococcus, and/or others which are a regular part of the urethral or intestinal, but not vaginal flora. In non-specific vaginitis, normal Lactobacillus flora is reduced or absent. Gardnerella vaginalis is considered to be the indicator organism for non-specific vaginitis which, in fact, is a polymicrobial infection.3,4 Although non- culture methods such as a direct Gram stain have been recommended in recent years for genital specimens, culture is still preferred by many laboratories.1,5 G. vaginalis may also be responsible for a variety of other diseases such as preterm birth, chorioamnionitis, urinary tract infections, newborn infections, and septicemia.6 The detection of the organism on routinely used media is difficult since Gardnerella and other -

MIB–MIP Is a Mycoplasma System That Captures and Cleaves Immunoglobulin G
MIB–MIP is a mycoplasma system that captures and cleaves immunoglobulin G Yonathan Arfia,b,1, Laetitia Minderc,d, Carmelo Di Primoe,f,g, Aline Le Royh,i,j, Christine Ebelh,i,j, Laurent Coquetk, Stephane Claveroll, Sanjay Vasheem, Joerg Joresn,o, Alain Blancharda,b, and Pascal Sirand-Pugneta,b aINRA (Institut National de la Recherche Agronomique), UMR 1332 Biologie du Fruit et Pathologie, F-33882 Villenave d’Ornon, France; bUniversity of Bordeaux, UMR 1332 Biologie du Fruit et Pathologie, F-33882 Villenave d’Ornon, France; cInstitut Européen de Chimie et Biologie, UMS 3033, University of Bordeaux, 33607 Pessac, France; dInstitut Bergonié, SIRIC BRIO, 33076 Bordeaux, France; eINSERM U1212, ARN Regulation Naturelle et Artificielle, 33607 Pessac, France; fCNRS UMR 5320, ARN Regulation Naturelle et Artificielle, 33607 Pessac, France; gInstitut Européen de Chimie et Biologie, University of Bordeaux, 33607 Pessac, France; hInstitut de Biologie Structurale, University of Grenoble Alpes, F-38044 Grenoble, France; iCNRS, Institut de Biologie Structurale, F-38044 Grenoble, France; jCEA, Institut de Biologie Structurale, F-38044 Grenoble, France; kCNRS UMR 6270, Plateforme PISSARO, Institute for Research and Innovation in Biomedicine - Normandie Rouen, Normandie Université, F-76821 Mont-Saint-Aignan, France; lProteome Platform, Functional Genomic Center of Bordeaux, University of Bordeaux, F-33076 Bordeaux Cedex, France; mJ. Craig Venter Institute, Rockville, MD 20850; nInternational Livestock Research Institute, 00100 Nairobi, Kenya; and oInstitute of Veterinary Bacteriology, University of Bern, CH-3001 Bern, Switzerland Edited by Roy Curtiss III, University of Florida, Gainesville, FL, and approved March 30, 2016 (received for review January 12, 2016) Mycoplasmas are “minimal” bacteria able to infect humans, wildlife, introduced into naive herds (8). -

Genomic Islands in Mycoplasmas
G C A T T A C G G C A T genes Review Genomic Islands in Mycoplasmas Christine Citti * , Eric Baranowski * , Emilie Dordet-Frisoni, Marion Faucher and Laurent-Xavier Nouvel Interactions Hôtes-Agents Pathogènes (IHAP), Université de Toulouse, INRAE, ENVT, 31300 Toulouse, France; [email protected] (E.D.-F.); [email protected] (M.F.); [email protected] (L.-X.N.) * Correspondence: [email protected] (C.C.); [email protected] (E.B.) Received: 30 June 2020; Accepted: 20 July 2020; Published: 22 July 2020 Abstract: Bacteria of the Mycoplasma genus are characterized by the lack of a cell-wall, the use of UGA as tryptophan codon instead of a universal stop, and their simplified metabolic pathways. Most of these features are due to the small-size and limited-content of their genomes (580–1840 Kbp; 482–2050 CDS). Yet, the Mycoplasma genus encompasses over 200 species living in close contact with a wide range of animal hosts and man. These include pathogens, pathobionts, or commensals that have retained the full capacity to synthesize DNA, RNA, and all proteins required to sustain a parasitic life-style, with most being able to grow under laboratory conditions without host cells. Over the last 10 years, comparative genome analyses of multiple species and strains unveiled some of the dynamics of mycoplasma genomes. This review summarizes our current knowledge of genomic islands (GIs) found in mycoplasmas, with a focus on pathogenicity islands, integrative and conjugative elements (ICEs), and prophages. Here, we discuss how GIs contribute to the dynamics of mycoplasma genomes and how they participate in the evolution of these minimal organisms. -

Serological Evidence That Chlamydiae and Mycoplasmas Are Involved in Infertility of Women B
Serological evidence that chlamydiae and mycoplasmas are involved in infertility of women B. R. M\l=o/\ller,D. Taylor-Robinson, Patricia M. Furr, B. Toft and J. Allen Division of Sexually Transmitted Diseases, MRC Clinical Research Centre, Watford Road, Harrow, Middlesex HAI 3UJ, U.K., and ^Department of Obstetrics and Gynaecology, University of Aarhus, DK-8000, Aarhus, Denmark Summary. Women with a history of infertility for 2 or more years were examined by hysterosalpingography (HSG) and antibodies against Chlamydia trachomatis, Myco- plasma hominis and M. genitalium were measured by a microimmunofluorescence technique in sera obtained immediately before HSG. Of 45 women with abnormal HSG findings, 15 (33%) had antibodies to C. trachomatis and 16 (35\m=.\5%)to M. hominis. In contrast, of 61 women with normal HSG findings, only 8 (13%) and 7 (11\m=.\5%)had antibodies to these micro-organisms, respectively. Antibody against M. genitalium was found in 26 of the patients (20% abnormal HSG and 28% normal HSG), indicating the need for further investigation of the significance of this mycoplasma in female infertility. The present results do confirm, however, that C. trachomatis is an important cause of infertility in women and suggest strongly that M. hominis is implicated. Introduction Infertility in women is caused often by tubai damage after pelvic inflammatory disease. Chlamydia trachomatis is a well-known pathogen in upper genital-tract infections and accounts for 25-50% of all cases of pelvic inflammatory disease (Paavonen, 1979) while Mycoplasma hominis is believed to be responsible for about 25% of all the cases (Moller, 1983). -

Human Microbiota Network: Unveiling Potential Crosstalk Between the Different Microbiota Ecosystems and Their Role in Health and Disease
nutrients Review Human Microbiota Network: Unveiling Potential Crosstalk between the Different Microbiota Ecosystems and Their Role in Health and Disease Jose E. Martínez †, Augusto Vargas † , Tania Pérez-Sánchez , Ignacio J. Encío , Miriam Cabello-Olmo * and Miguel Barajas * Biochemistry Area, Department of Health Science, Public University of Navarre, 31008 Pamplona, Spain; [email protected] (J.E.M.); [email protected] (A.V.); [email protected] (T.P.-S.); [email protected] (I.J.E.) * Correspondence: [email protected] (M.C.-O.); [email protected] (M.B.) † These authors contributed equally to this work. Abstract: The human body is host to a large number of microorganisms which conform the human microbiota, that is known to play an important role in health and disease. Although most of the microorganisms that coexist with us are located in the gut, microbial cells present in other locations (like skin, respiratory tract, genitourinary tract, and the vaginal zone in women) also play a significant role regulating host health. The fact that there are different kinds of microbiota in different body areas does not mean they are independent. It is plausible that connection exist, and different studies have shown that the microbiota present in different zones of the human body has the capability of communicating through secondary metabolites. In this sense, dysbiosis in one body compartment Citation: Martínez, J.E.; Vargas, A.; may negatively affect distal areas and contribute to the development of diseases. Accordingly, it Pérez-Sánchez, T.; Encío, I.J.; could be hypothesized that the whole set of microbial cells that inhabit the human body form a Cabello-Olmo, M.; Barajas, M. -
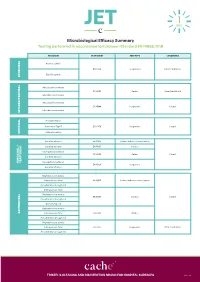
JET Microbiological Efficacy Summary
Microbiological Efficacy Summary Testing performed in accordance to European Standard EN 14885:2018 ORGANISM TEST NORM TEST TYPE CONDITIONS Bacillus subtilis EN 13704 Suspension Clean 1 and Dirty 1 Bacillus cereus SPORICIDAL Mycobacterium terrae EN 14563 Carrier Clean 1 and Dirty 2 Mycobacterium avium Mycobacterium terrae EN 14348 Suspension Clean 1 Mycobacterium avium MYCOBACTERICIDAL Poliovirus Type 1 Adenovirus Type 5 EN 14476 Suspension Clean 1 Murine Norovirus VIRUCIDAL Candida albicans EN 16615 Surface with mechanical action Candida albicans EN 13697 Surface Aspergillus brasiliensis EN 14562 Carrier Clean 1 Candida albicans YEASTICIDAL FUNGICIDAL / FUNGICIDAL Aspergillus brasiliensis EN 13624 Suspension Candida albicans Staphylococcus aureus Enterococcus hirae EN 16615 Surface with mechanical action Pseudomonas aeruginosa Enterococcus hirae Staphylococcus aureus EN 13697 Surface Clean 1 Pseudomonas aeruginosa Escherichia coli Staphylococcus aureus BACTERICIDAL Enterococcus hirae EN 14561 Carrier Pseudomonas aeruginosa Staphylococcus aureus Enterococcus hirae EN 13727 Suspension Clean 1 and Dirty 1 Pseudomonas aeruginosa TRISTEL’S CLEANING AND DISINFECTION BRAND FOR HOSPITAL SURFACES Page 1 of 3 Additional Testing TEST METHOD RNA DNA / Polyacrylamide gel electrophoresis (PAGE) ORGANISM TEST METHOD TEST TYPE CONDITIONS Acanthamoeba castellanii cysts Following the method of EN 13704 Suspension Clean 1 PROTOZOA Bacillus subtilis EN 17126 Suspension Clean 1 Bacillus cereus Clostridium difficile EN 13704 Suspension Clean 1 and Dirty 1 -

NOTES in Vitro Activities of Norfloxacin and Ciprofloxacin Against
ANTIMICROBIAL AGENTS AND CHEMOTHERAPY, July 1984, p. 94-96 Vol. 26, No. 1 0066-4804/84/070094-03$02.00/0 Copyright C 1984, American Society for Microbiology NOTES In Vitro Activities of Norfloxacin and Ciprofloxacin Against Mycobacterium tuberculosis, M. avium Complex, M. chelonei, M. fortuitum, and M. kansasii J. DOUGLAS GAY, DONALD R. DEYOUNG, AND GLENN D. ROBERTS* Section of Clinical Microbiology, Department of Laboratory Medicine, Mayo Clinic and Mayo Foundation, Rochester, Minnesota 55905 Received 28 November 1983/Accepted 4 April 1984 The activities of ciprofloxacin and norfloxacin against 100 mycobacteria isolates were studied in vitro by the 1% standard proportion method. Ciprofloxacin was more active against M. tuberculosis and M. fortuitum with MICs of 1.0 and 0.25 ,ug/ml, respectively, against 90% of isolates; norfloxacin had MICs of 8.0 and 2.0 ,ug/ml, respectively, against 90% of isolates. Nalidixic acid and other heterocyclic carbonic acid deriva- studied. The organisms were taken from the Mayo Clinic tives have been used primarily in the treatment of urinary stock culture collection, which included recent clinical iso- tract infections for many years. The compounds of this lates. Stock cultures were maintained on Middlebrook 7H10 general group include nalidixic acid, oxolinic acid, pipemidic agar slants (Difco Laboratories, Detroit, Mich.) and were acid, cinoxacin, and rosoxacin. Two new substances in this subcultured monthly. The identification of isolates was series which have been recently synthesized are norfloxacin based on standard biochemical tests (17) and gas-liquid (6) (1-ethyl-6-fluoro-1,4-dihydro-4-oxo-7-[ 1-piperazinyl ]-3- chromatography (16). -

A Genomic Journey Through a Genus of Large DNA Viruses
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Virology Papers Virology, Nebraska Center for 2013 Towards defining the chloroviruses: a genomic journey through a genus of large DNA viruses Adrien Jeanniard Aix-Marseille Université David D. Dunigan University of Nebraska-Lincoln, [email protected] James Gurnon University of Nebraska-Lincoln, [email protected] Irina V. Agarkova University of Nebraska-Lincoln, [email protected] Ming Kang University of Nebraska-Lincoln, [email protected] See next page for additional authors Follow this and additional works at: https://digitalcommons.unl.edu/virologypub Part of the Biological Phenomena, Cell Phenomena, and Immunity Commons, Cell and Developmental Biology Commons, Genetics and Genomics Commons, Infectious Disease Commons, Medical Immunology Commons, Medical Pathology Commons, and the Virology Commons Jeanniard, Adrien; Dunigan, David D.; Gurnon, James; Agarkova, Irina V.; Kang, Ming; Vitek, Jason; Duncan, Garry; McClung, O William; Larsen, Megan; Claverie, Jean-Michel; Van Etten, James L.; and Blanc, Guillaume, "Towards defining the chloroviruses: a genomic journey through a genus of large DNA viruses" (2013). Virology Papers. 245. https://digitalcommons.unl.edu/virologypub/245 This Article is brought to you for free and open access by the Virology, Nebraska Center for at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Virology Papers by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Authors Adrien Jeanniard, David D. Dunigan, James Gurnon, Irina V. Agarkova, Ming Kang, Jason Vitek, Garry Duncan, O William McClung, Megan Larsen, Jean-Michel Claverie, James L. Van Etten, and Guillaume Blanc This article is available at DigitalCommons@University of Nebraska - Lincoln: https://digitalcommons.unl.edu/ virologypub/245 Jeanniard, Dunigan, Gurnon, Agarkova, Kang, Vitek, Duncan, McClung, Larsen, Claverie, Van Etten & Blanc in BMC Genomics (2013) 14. -

Sample Report Vaginosis.Pdf
LAB #: Sample Report CLIENT #: 12345 PATIENT: Sample Patient DOCTOR: Sample Doctor ID: Doctor's Data, Inc. SEX: Female 3755 Illinois Ave. DOB: 01/01/1993 AGE: 25 St. Charles, IL 60174 U.S.A. !"#$%&'(%(!)*'+%,- GRAM STAIN MICROSCOPY BACTERIAL VAGINOSIS SCORE Normal Abnormal Expected score interpretation: Lactobacilli None Mod - Many 0 - 3 BV not likely 4 - 6 BV indeterminate Curved Gram 10 7-10 BV highly suggestive Negative Rods Many None Small Gram 1 Many None The BV score is calculated based upon the gram stain results Negative Rods and is independent of the yeast, and bacterial cultures. 1Nugent Scoring System. (Nugent et al. J. Clin. Micro. Yeast None None (1991)29:297-301) RBC’s None None YEAST CULTURE 2+ Candida albicans WBC’s 0 0 - 6 Clue Cells Present None Eosinophils N/A None Eosinophils reported and Wrights Stain performed when WBC’s >6 Additional Gram Stain Findings: Rare Gram positive cocci in pairs BACTERIOLOGY CULTURE Expected/Beneficial flora Commensal (Imbalanced) flora Dysbiotic flora NG Lactobacillus spp. 2+ Alpha hemolytic strep 3+ Gardnerella vaginalis 2+ Gamma hemolytic strep 4+ Enterococcus faecalis 1+ Staphylococcus not aureus NG = No Growth SPECIMEN DATA Comments: Date Collected: 01/14/2019 Date Received: 01/16/2019 Date Completed: 01/23/2019 ©Doctor’s Data, Inc. !!! ADDRESS: 3755 Illinois Avenue, St. Charles, IL 60174-2420 !!! MED DIR: Erlo Roth, MD !!! CLIA ID NO: 14D0646470 0002038 LAB #: Sample Report CLIENT #: 12345 PATIENT: Sample DOCTOR: Sample Doctor Patient ID: Doctor's Data, Inc. SEX: Female 3755 Illinois Ave. DOB: 01/01/1993 St. Charles, IL 60174 U.S.A. -

Mycobacterium Avium Possesses Extracellular DNA That Contributes to Biofilm Formation, Structural Integrity, and Tolerance to Antibiotics
Mycobacterium avium Possesses Extracellular DNA that Contributes to Biofilm Formation, Structural Integrity, and Tolerance to Antibiotics Rose, S. J., Babrak, L. M., & Bermudez, L. E. (2015). Mycobacterium avium Possesses Extracellular DNA that Contributes to Biofilm Formation, Structural Integrity, and Tolerance to Antibiotics. PLoS ONE, 10(5), e0128772. doi: 10.1371/journal.pone.0128772 10.1371/journal.pone.0128772 Public Library of Science Version of Record http://cdss.library.oregonstate.edu/sa-termsofuse RESEARCH ARTICLE Mycobacterium avium Possesses Extracellular DNA that Contributes to Biofilm Formation, Structural Integrity, and Tolerance to Antibiotics Sasha J. Rose1,2, Lmar M. Babrak1,2, Luiz E. Bermudez1,2* 1 Department of Biomedical Sciences, College of Veterinary Medicine, Oregon State University, Corvallis, Oregon, United States of America, 2 Department of Microbiology, College of Science, Oregon State University, Corvallis, Oregon, United States of America * [email protected] Abstract Mycobacterium avium subsp. hominissuis is an opportunistic pathogen that is associated with biofilm-related infections of the respiratory tract and is difficult to treat. In recent years, extracellular DNA (eDNA) has been found to be a major component of bacterial biofilms, in- OPEN ACCESS cluding many pathogens involved in biofilm-associated infections. To date, eDNA has not Citation: Rose SJ, Babrak LM, Bermudez LE (2015) been described as a component of mycobacterial biofilms. In this study, we identified and Mycobacterium avium Possesses Extracellular DNA characterized eDNA in a high biofilm-producing strain of Mycobacterium avium subsp. that Contributes to Biofilm Formation, Structural hominissuis (MAH). In addition, we surveyed for presence of eDNA in various MAH strains Integrity, and Tolerance to Antibiotics. -

Prevalence of Ureaplasma Urealyticum, Mycoplasma Hominis and Chlamydia Trachomatis in Patients with Uncomplicated Recurrent Urin
Nephrology and Renal Diseases Research Article ISSN: 2399-908X Prevalence of Ureaplasma urealyticum, Mycoplasma hominis and Chlamydia trachomatis in patients with uncomplicated recurrent urinary tract infections Jadranka Vlasic-Matas1*, Hrvoje Raos2, Marijana Vuckovic2, Stjepan Radic2 and Vesna Capkun3 1Polyclinic Nephrology Department, Split, Croatia 2School of Medicine, University of Split, Split, Croatia 3Department of Nuclear Medicine, Split University Hospital Center, Split, Croatia Abstract Aim: To assess the prevalence of Ureaplasma urealyticum, Mycoplasma hominis and Chlamydia trachomatis in patients with chronic urinary tract infections (UTIs) and its correlation with leukocyturia and symptoms. Methods: The study included 220 patients (130 women and 90 men) presenting with chronic voiding symptoms and sterile leukocyturia. Urine, urethral swabs and cervical swabs (for women patients) were taken to determine the presence of these pathogens. Patients were treated by tetracycline and followed up three and six months after initial therapy. Results: In 186 (85%) out of 220 patients, U. urealyticum was found, while C. trachomatis was present in 34 patients (15%). In majority of female patients (112 out of 130; 86%) U. urealyticum was found. In addition to ureaplasma, in eight patients M. hominis was found. C. trachomatis was identified in 18 female patients (14%). In 74 out of 90 (82%) male patients U. urealyticum was detected while in six of them M. hominis was also found. C. trachomatis was identified in 16 male patients (18%). U. urealyticum was significantly related to leukocyturia, as opposed to C. trachomatis (p<0,001). Women had more frequent symptomatology (p = 0,015) and higer leukocyturia (p<0.001). Conclusion: Leukocyturia is more common find in U. -

Characterization of an Α-Glucosidase Enzyme Conserved in Gardnerella
bioRxiv preprint doi: https://doi.org/10.1101/2020.05.11.086124; this version posted May 11, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Characterization of an a-glucosidase enzyme conserved in Gardnerella spp. isolated 2 from the human vaginal microbiome 3 4 Pashupati Bhandari1, Jeffrey P. Tingley2, D. Wade Abbott2 and Janet E. Hill1,* 5 6 1Department of Veterinary Microbiology, Western College of Veterinary Medicine, 7 University of Saskatchewan, 52 Campus Drive, Saskatoon, Saskatchewan, S7N 5B4, 8 Canada 9 2Lethbridge Research and Development Centre, Agriculture and Agri-Food Canada, 10 Lethbridge, Alberta, T1J 4B1, Canada 11 12 *To whom correspondence should be addressed 13 [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.05.11.086124; this version posted May 11, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 14 Abstract 15 Gardnerella spp. in the vaginal microbiome are associated with bacterial vaginosis, a 16 dysbiosis in which lactobacilli dominant microbial community is replaced with mixed 17 aerobic and anaerobic bacteria including Gardnerella species. The co-occurrence of 18 multiple Gardnerella species in the vaginal environment is common, but different species 19 are dominant in different women.