Examining the Link Between Macrophyte Diversity, Bacterial
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Chemical Structures of Some Examples of Earlier Characterized Antibiotic and Anticancer Specialized
Supplementary figure S1: Chemical structures of some examples of earlier characterized antibiotic and anticancer specialized metabolites: (A) salinilactam, (B) lactocillin, (C) streptochlorin, (D) abyssomicin C and (E) salinosporamide K. Figure S2. Heat map representing hierarchical classification of the SMGCs detected in all the metagenomes in the dataset. Table S1: The sampling locations of each of the sites in the dataset. Sample Sample Bio-project Site depth accession accession Samples Latitude Longitude Site description (m) number in SRA number in SRA AT0050m01B1-4C1 SRS598124 PRJNA193416 Atlantis II water column 50, 200, Water column AT0200m01C1-4D1 SRS598125 21°36'19.0" 38°12'09.0 700 and above the brine N "E (ATII 50, ATII 200, 1500 pool water layers AT0700m01C1-3D1 SRS598128 ATII 700, ATII 1500) AT1500m01B1-3C1 SRS598129 ATBRUCL SRS1029632 PRJNA193416 Atlantis II brine 21°36'19.0" 38°12'09.0 1996– Brine pool water ATBRLCL1-3 SRS1029579 (ATII UCL, ATII INF, N "E 2025 layers ATII LCL) ATBRINP SRS481323 PRJNA219363 ATIID-1a SRS1120041 PRJNA299097 ATIID-1b SRS1120130 ATIID-2 SRS1120133 2168 + Sea sediments Atlantis II - sediments 21°36'19.0" 38°12'09.0 ~3.5 core underlying ATII ATIID-3 SRS1120134 (ATII SDM) N "E length brine pool ATIID-4 SRS1120135 ATIID-5 SRS1120142 ATIID-6 SRS1120143 Discovery Deep brine DDBRINP SRS481325 PRJNA219363 21°17'11.0" 38°17'14.0 2026– Brine pool water N "E 2042 layers (DD INF, DD BR) DDBRINE DD-1 SRS1120158 PRJNA299097 DD-2 SRS1120203 DD-3 SRS1120205 Discovery Deep 2180 + Sea sediments sediments 21°17'11.0" -

Catalogue of Bacteria Shapes
We first tried to use the most general shape associated with each genus, which are often consistent across species (spp.) (first choice for shape). If there was documented species variability, either the most common species (second choice for shape) or well known species (third choice for shape) is shown. Corynebacterium: pleomorphic bacilli. Due to their snapping type of division, cells often lie in clusters resembling chinese letters (https://microbewiki.kenyon.edu/index.php/Corynebacterium) Shown is Corynebacterium diphtheriae Figure 1. Stained Corynebacterium cells. The "barred" appearance is due to the presence of polyphosphate inclusions called metachromatic granules. Note also the characteristic "Chinese-letter" arrangement of cells. (http:// textbookofbacteriology.net/diphtheria.html) Lactobacillus: Lactobacilli are rod-shaped, Gram-positive, fermentative, organotrophs. They are usually straight, although they can form spiral or coccobacillary forms under certain conditions. (https://microbewiki.kenyon.edu/index.php/ Lactobacillus) Porphyromonas: A genus of small anaerobic gram-negative nonmotile cocci and usually short rods thatproduce smooth, gray to black pigmented colonies the size of which varies with the species. (http:// medical-dictionary.thefreedictionary.com/Porphyromonas) Shown: Porphyromonas gingivalis Moraxella: Moraxella is a genus of Gram-negative bacteria in the Moraxellaceae family. It is named after the Swiss ophthalmologist Victor Morax. The organisms are short rods, coccobacilli or, as in the case of Moraxella catarrhalis, diplococci in morphology (https://en.wikipedia.org/wiki/Moraxella). *This one could be changed to a diplococcus shape because of moraxella catarrhalis, but i think the short rods are fair given the number of other moraxella with them. Jeotgalicoccus: Jeotgalicoccus is a genus of Gram-positive, facultatively anaerobic, and halotolerant to halophilicbacteria. -

BD™ Gardnerella Selective Agar with 5% Human Blood
INSTRUCTIONS FOR USE – READY-TO-USE PLATED MEDIA PA-254094.06 Rev.: July 2014 BD Gardnerella Selective Agar with 5% Human Blood INTENDED USE BD Gardnerella Selective Agar with 5% Human Blood is a partially selective and differential medium for the isolation of Gardnerella vaginalis from clinical specimens. PRINCIPLES AND EXPLANATION OF THE PROCEDURE Microbiological method. Gardnerella vaginalis is considered to be one of the organisms causing vaginitis.1-4 Although the organism may be present in a high percentage of normal women in the vaginal flora, its importance as a cause of non-specific vaginitis (also called bacterial vaginosis) has never been questioned. In symptomatic women, G. vaginalis frequently is associated with anaerobes such as Prevotella bivia, P. disiens, Mobiluncus, Peptostreptococcus, and/or others which are a regular part of the urethral or intestinal, but not vaginal flora. In non-specific vaginitis, normal Lactobacillus flora is reduced or absent. Gardnerella vaginalis is considered to be the indicator organism for non-specific vaginitis which, in fact, is a polymicrobial infection.3,4 Although non- culture methods such as a direct Gram stain have been recommended in recent years for genital specimens, culture is still preferred by many laboratories.1,5 G. vaginalis may also be responsible for a variety of other diseases such as preterm birth, chorioamnionitis, urinary tract infections, newborn infections, and septicemia.6 The detection of the organism on routinely used media is difficult since Gardnerella and other -

MIB–MIP Is a Mycoplasma System That Captures and Cleaves Immunoglobulin G
MIB–MIP is a mycoplasma system that captures and cleaves immunoglobulin G Yonathan Arfia,b,1, Laetitia Minderc,d, Carmelo Di Primoe,f,g, Aline Le Royh,i,j, Christine Ebelh,i,j, Laurent Coquetk, Stephane Claveroll, Sanjay Vasheem, Joerg Joresn,o, Alain Blancharda,b, and Pascal Sirand-Pugneta,b aINRA (Institut National de la Recherche Agronomique), UMR 1332 Biologie du Fruit et Pathologie, F-33882 Villenave d’Ornon, France; bUniversity of Bordeaux, UMR 1332 Biologie du Fruit et Pathologie, F-33882 Villenave d’Ornon, France; cInstitut Européen de Chimie et Biologie, UMS 3033, University of Bordeaux, 33607 Pessac, France; dInstitut Bergonié, SIRIC BRIO, 33076 Bordeaux, France; eINSERM U1212, ARN Regulation Naturelle et Artificielle, 33607 Pessac, France; fCNRS UMR 5320, ARN Regulation Naturelle et Artificielle, 33607 Pessac, France; gInstitut Européen de Chimie et Biologie, University of Bordeaux, 33607 Pessac, France; hInstitut de Biologie Structurale, University of Grenoble Alpes, F-38044 Grenoble, France; iCNRS, Institut de Biologie Structurale, F-38044 Grenoble, France; jCEA, Institut de Biologie Structurale, F-38044 Grenoble, France; kCNRS UMR 6270, Plateforme PISSARO, Institute for Research and Innovation in Biomedicine - Normandie Rouen, Normandie Université, F-76821 Mont-Saint-Aignan, France; lProteome Platform, Functional Genomic Center of Bordeaux, University of Bordeaux, F-33076 Bordeaux Cedex, France; mJ. Craig Venter Institute, Rockville, MD 20850; nInternational Livestock Research Institute, 00100 Nairobi, Kenya; and oInstitute of Veterinary Bacteriology, University of Bern, CH-3001 Bern, Switzerland Edited by Roy Curtiss III, University of Florida, Gainesville, FL, and approved March 30, 2016 (received for review January 12, 2016) Mycoplasmas are “minimal” bacteria able to infect humans, wildlife, introduced into naive herds (8). -

CALIFORNIA STATE UNIVERSITY, NORTHRIDGE Comparative
CALIFORNIA STATE UNIVERSITY, NORTHRIDGE Comparative Genomics and Epigenomics of Sporosarcina ureae A thesis submitted in partial fulfillment of the requirement for the degree of Master of Science in Biology By Andrew Oliver August 2016 The thesis of Andrew Oliver is approved by: _________________________________________ ____________ Sean Murray, Ph.D. Date _________________________________________ ____________ Gilberto Flores, Ph.D. Date _________________________________________ ____________ Kerry Cooper, Ph.D., Chair Date California State University, Northridge ii Acknowledgments First and foremost, a special thanks to my advisor, Dr. Kerry Cooper, for his advice and, above all, his patience. If I can be half the scientist you are someday, I would be thrilled. I would like to also thank everyone in the Cooper lab, especially my colleagues Courtney Sams and Tabitha Bayangnos. It was a privilege to work along side you. More thanks to my committee members, Dr. Gilberto Flores and Dr. Sean Murray. Dr. Flores, you were instrumental in guiding me to ask the right questions regarding bacterial taxonomy. Dr. Murray, your contributions to my graduate studies would make this section run on for pages. I thank you for taking me under your wing from the beginning. Acknowledgement and thanks to the Baresi lab, especially Dr. Larry Baresi and Tania Kurbessoian for their partnership in this research. Also to Bernardine Pregerson for all the work that lays at the foundation of this study. This research would not be what it is without the help of my childhood friend, Matthew Kay. You wrote programs, taught me coding languages, and challenged me to go digging for answers to very difficult questions. -

Table S5. the Information of the Bacteria Annotated in the Soil Community at Species Level
Table S5. The information of the bacteria annotated in the soil community at species level No. Phylum Class Order Family Genus Species The number of contigs Abundance(%) 1 Firmicutes Bacilli Bacillales Bacillaceae Bacillus Bacillus cereus 1749 5.145782459 2 Bacteroidetes Cytophagia Cytophagales Hymenobacteraceae Hymenobacter Hymenobacter sedentarius 1538 4.52499338 3 Gemmatimonadetes Gemmatimonadetes Gemmatimonadales Gemmatimonadaceae Gemmatirosa Gemmatirosa kalamazoonesis 1020 3.000970902 4 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas indica 797 2.344876284 5 Firmicutes Bacilli Lactobacillales Streptococcaceae Lactococcus Lactococcus piscium 542 1.594633558 6 Actinobacteria Thermoleophilia Solirubrobacterales Conexibacteraceae Conexibacter Conexibacter woesei 471 1.385742446 7 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas taxi 430 1.265115184 8 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas wittichii 388 1.141545794 9 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas sp. FARSPH 298 0.876754244 10 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sorangium cellulosum 260 0.764953367 11 Proteobacteria Deltaproteobacteria Myxococcales Polyangiaceae Sorangium Sphingomonas sp. Cra20 260 0.764953367 12 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas panacis 252 0.741416341 -

Genomic Islands in Mycoplasmas
G C A T T A C G G C A T genes Review Genomic Islands in Mycoplasmas Christine Citti * , Eric Baranowski * , Emilie Dordet-Frisoni, Marion Faucher and Laurent-Xavier Nouvel Interactions Hôtes-Agents Pathogènes (IHAP), Université de Toulouse, INRAE, ENVT, 31300 Toulouse, France; [email protected] (E.D.-F.); [email protected] (M.F.); [email protected] (L.-X.N.) * Correspondence: [email protected] (C.C.); [email protected] (E.B.) Received: 30 June 2020; Accepted: 20 July 2020; Published: 22 July 2020 Abstract: Bacteria of the Mycoplasma genus are characterized by the lack of a cell-wall, the use of UGA as tryptophan codon instead of a universal stop, and their simplified metabolic pathways. Most of these features are due to the small-size and limited-content of their genomes (580–1840 Kbp; 482–2050 CDS). Yet, the Mycoplasma genus encompasses over 200 species living in close contact with a wide range of animal hosts and man. These include pathogens, pathobionts, or commensals that have retained the full capacity to synthesize DNA, RNA, and all proteins required to sustain a parasitic life-style, with most being able to grow under laboratory conditions without host cells. Over the last 10 years, comparative genome analyses of multiple species and strains unveiled some of the dynamics of mycoplasma genomes. This review summarizes our current knowledge of genomic islands (GIs) found in mycoplasmas, with a focus on pathogenicity islands, integrative and conjugative elements (ICEs), and prophages. Here, we discuss how GIs contribute to the dynamics of mycoplasma genomes and how they participate in the evolution of these minimal organisms. -

Sporosarcina Aquimarina Sjam16103 Isolated from the Pneumatophores of Avicennia Marina L
Hindawi Publishing Corporation International Journal of Microbiology Volume 2012, Article ID 532060, 10 pages doi:10.1155/2012/532060 Research Article Plant Growth Promoting of Endophytic Sporosarcina aquimarina SjAM16103 Isolated from the Pneumatophores of Avicennia marina L. S. Rylo Sona Janarthine1 and P. Eganathan2 1 Faculty of Marine Science, Annamalai University, Chidambaram 608 502, India 2 Biotechnology Division, M S Swaminathan Research Foundation, Chennai 600 113, India Correspondence should be addressed to S. Rylo Sona Janarthine, jana [email protected] Received 17 October 2011; Revised 12 January 2012; Accepted 20 April 2012 AcademicEditor:A.J.M.Stams Copyright © 2012 S. R. S. Janarthine and P. Eganathan. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Endophytic Sporosarcina aquimarina SjAM16103 was isolated from the inner tissues of pneumatophores of mangrove plant Avicennia marina along with Bacillus sp. and Enterobacter sp. Endophytic S. aquimarina SjAM16103 was Gram variable, and motile bacterium measured 0.6–0.9 μm wide by 1.7–2.0 μm long and light orange-brown coloured in 3-day cultures on tryptone broth at 26◦C. Nucleotide sequence of this strain has been deposited in the GenBank under accession number GU930359. This endophytic bacterium produced 2.37 μMol/mL of indole acetic acid and siderophore as it metabolites. This strain could solubilize phosphate molecules and fixes atmospheric nitrogen. Endophytic S. aquimarina SjAM16103 was inoculated into four different plants under in vitro method to analyse its growth-promoting activity and role inside the host plants. -

The Power of Partnership » AP2.Com MOLECULAR TEST MENU
The Power of Partnership » AP2.com MOLECULAR TEST MENU AP2 provides the most comprehensive Women’s Health Care molecular menu. AP2 can perform most of these tests from our scientifically developed UniSwabTM as well as from the liquid based platforms SurePathTM and ThinPrep®. The following is a comprehensive list of molecular tests available from AP2. If one molecular test is ordered upfront, add-on testing is available for 45 days on UniSwabTM, ThinPrep® and SurePathTM. If no molecular test is ordered up front, add-on testing is available for 28 days on UniSwabTM and ThinPrep®. HPV and CT/NG testing can be added on to ThinPrep® specimens prior to 28 days after collection. UniSwab ThinPrep SurePath 70142 Atopobium vaginae 70142 Atopobium vaginae 70142 Atopobium vaginae 60135 Bacterial vaginosis panel 60135 Bacterial vaginosis panel 60135 Bacterial vaginosis panel (Mobiluncus mulieris and M. curtisii, Gardnerella (Mobiluncus mulieris and M. curtisii, Gardnerella (Mobiluncus mulieris and M. curtisii, Gardnerella vaginalis, Atopobium vaginae, Mycoplasma vaginalis, Atopobium vaginae, Mycoplasma vaginalis, Atopobium vaginae, Mycoplasma genitalium and hominis) genitalium and hominis) genitalium and hominis) 70125 Bacteroides fragilis 70125 Bacteroides fragilis 70125 Bacteroides fragilis 70164 BVAB2 70164 BVAB2 70164 BVAB2 70551 Candida albicans 70551 Candida albicans 70551 Candida albicans 70559 Candida glabrata 70559 Candida glabrata 70559 Candida glabrata 70561 Candida kefyr 70561 Candida kefyr 70561 Candida kefyr 70560 Candida krusei 70560 -

Serological Evidence That Chlamydiae and Mycoplasmas Are Involved in Infertility of Women B
Serological evidence that chlamydiae and mycoplasmas are involved in infertility of women B. R. M\l=o/\ller,D. Taylor-Robinson, Patricia M. Furr, B. Toft and J. Allen Division of Sexually Transmitted Diseases, MRC Clinical Research Centre, Watford Road, Harrow, Middlesex HAI 3UJ, U.K., and ^Department of Obstetrics and Gynaecology, University of Aarhus, DK-8000, Aarhus, Denmark Summary. Women with a history of infertility for 2 or more years were examined by hysterosalpingography (HSG) and antibodies against Chlamydia trachomatis, Myco- plasma hominis and M. genitalium were measured by a microimmunofluorescence technique in sera obtained immediately before HSG. Of 45 women with abnormal HSG findings, 15 (33%) had antibodies to C. trachomatis and 16 (35\m=.\5%)to M. hominis. In contrast, of 61 women with normal HSG findings, only 8 (13%) and 7 (11\m=.\5%)had antibodies to these micro-organisms, respectively. Antibody against M. genitalium was found in 26 of the patients (20% abnormal HSG and 28% normal HSG), indicating the need for further investigation of the significance of this mycoplasma in female infertility. The present results do confirm, however, that C. trachomatis is an important cause of infertility in women and suggest strongly that M. hominis is implicated. Introduction Infertility in women is caused often by tubai damage after pelvic inflammatory disease. Chlamydia trachomatis is a well-known pathogen in upper genital-tract infections and accounts for 25-50% of all cases of pelvic inflammatory disease (Paavonen, 1979) while Mycoplasma hominis is believed to be responsible for about 25% of all the cases (Moller, 1983). -

Human Microbiota Network: Unveiling Potential Crosstalk Between the Different Microbiota Ecosystems and Their Role in Health and Disease
nutrients Review Human Microbiota Network: Unveiling Potential Crosstalk between the Different Microbiota Ecosystems and Their Role in Health and Disease Jose E. Martínez †, Augusto Vargas † , Tania Pérez-Sánchez , Ignacio J. Encío , Miriam Cabello-Olmo * and Miguel Barajas * Biochemistry Area, Department of Health Science, Public University of Navarre, 31008 Pamplona, Spain; [email protected] (J.E.M.); [email protected] (A.V.); [email protected] (T.P.-S.); [email protected] (I.J.E.) * Correspondence: [email protected] (M.C.-O.); [email protected] (M.B.) † These authors contributed equally to this work. Abstract: The human body is host to a large number of microorganisms which conform the human microbiota, that is known to play an important role in health and disease. Although most of the microorganisms that coexist with us are located in the gut, microbial cells present in other locations (like skin, respiratory tract, genitourinary tract, and the vaginal zone in women) also play a significant role regulating host health. The fact that there are different kinds of microbiota in different body areas does not mean they are independent. It is plausible that connection exist, and different studies have shown that the microbiota present in different zones of the human body has the capability of communicating through secondary metabolites. In this sense, dysbiosis in one body compartment Citation: Martínez, J.E.; Vargas, A.; may negatively affect distal areas and contribute to the development of diseases. Accordingly, it Pérez-Sánchez, T.; Encío, I.J.; could be hypothesized that the whole set of microbial cells that inhabit the human body form a Cabello-Olmo, M.; Barajas, M. -
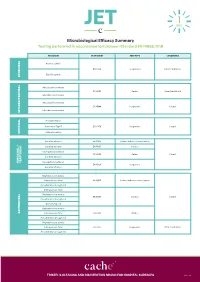
JET Microbiological Efficacy Summary
Microbiological Efficacy Summary Testing performed in accordance to European Standard EN 14885:2018 ORGANISM TEST NORM TEST TYPE CONDITIONS Bacillus subtilis EN 13704 Suspension Clean 1 and Dirty 1 Bacillus cereus SPORICIDAL Mycobacterium terrae EN 14563 Carrier Clean 1 and Dirty 2 Mycobacterium avium Mycobacterium terrae EN 14348 Suspension Clean 1 Mycobacterium avium MYCOBACTERICIDAL Poliovirus Type 1 Adenovirus Type 5 EN 14476 Suspension Clean 1 Murine Norovirus VIRUCIDAL Candida albicans EN 16615 Surface with mechanical action Candida albicans EN 13697 Surface Aspergillus brasiliensis EN 14562 Carrier Clean 1 Candida albicans YEASTICIDAL FUNGICIDAL / FUNGICIDAL Aspergillus brasiliensis EN 13624 Suspension Candida albicans Staphylococcus aureus Enterococcus hirae EN 16615 Surface with mechanical action Pseudomonas aeruginosa Enterococcus hirae Staphylococcus aureus EN 13697 Surface Clean 1 Pseudomonas aeruginosa Escherichia coli Staphylococcus aureus BACTERICIDAL Enterococcus hirae EN 14561 Carrier Pseudomonas aeruginosa Staphylococcus aureus Enterococcus hirae EN 13727 Suspension Clean 1 and Dirty 1 Pseudomonas aeruginosa TRISTEL’S CLEANING AND DISINFECTION BRAND FOR HOSPITAL SURFACES Page 1 of 3 Additional Testing TEST METHOD RNA DNA / Polyacrylamide gel electrophoresis (PAGE) ORGANISM TEST METHOD TEST TYPE CONDITIONS Acanthamoeba castellanii cysts Following the method of EN 13704 Suspension Clean 1 PROTOZOA Bacillus subtilis EN 17126 Suspension Clean 1 Bacillus cereus Clostridium difficile EN 13704 Suspension Clean 1 and Dirty 1