Prevalence of Ureaplasma Urealyticum, Mycoplasma Hominis and Chlamydia Trachomatis in Patients with Uncomplicated Recurrent Urin
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Comparative Genome Analysis of 19 Ureaplasma Urealyticum and Ureaplasma Parvum Strains
Paralanov et al. BMC Microbiology 2012, 12:88 http://www.biomedcentral.com/1471-2180/12/88 RESEARCH ARTICLE Open Access Comparative genome analysis of 19 Ureaplasma urealyticum and Ureaplasma parvum strains Vanya Paralanov1, Jin Lu2, Lynn B Duffy2, Donna M Crabb2, Susmita Shrivastava1, Barbara A Methé1, Jason Inman1, Shibu Yooseph1, Li Xiao2, Gail H Cassell2, Ken B Waites2 and John I Glass1* Abstract Background: Ureaplasma urealyticum (UUR) and Ureaplasma parvum (UPA) are sexually transmitted bacteria among humans implicated in a variety of disease states including but not limited to: nongonococcal urethritis, infertility, adverse pregnancy outcomes, chorioamnionitis, and bronchopulmonary dysplasia in neonates. There are 10 distinct serotypes of UUR and 4 of UPA. Efforts to determine whether difference in pathogenic potential exists at the ureaplasma serovar level have been hampered by limitations of antibody-based typing methods, multiple cross-reactions and poor discriminating capacity in clinical samples containing two or more serovars. Results: We determined the genome sequences of the American Type Culture Collection (ATCC) type strains of all UUR and UPA serovars as well as four clinical isolates of UUR for which we were not able to determine serovar designation. UPA serovars had 0.75−0.78 Mbp genomes and UUR serovars were 0.84−0.95 Mbp. The original classification of ureaplasma isolates into distinct serovars was largely based on differences in the major ureaplasma surface antigen called the multiple banded antigen (MBA) and reactions of human and animal sera to the organisms. Whole genome analysis of the 14 serovars and the 4 clinical isolates showed the mba gene was part of a large superfamily, which is a phase variable gene system, and that some serovars have identical sets of mba genes. -

Roles of Oral Bacteria in Cardiovascular Diseases—From
J Pharmacol Sci 113, 000 – 000 (2010) Journal of Pharmacological Sciences ©2010 The Japanese Pharmacological Society Forum Minireview Roles of Oral Bacteria in Cardiovascular Diseases — From Molecular Mechanisms to Clinical Cases: Implication of Periodontal Diseases in Development of Systemic Diseases Hiroaki Inaba1 and Atsuo Amano1,* 1Department of Oral Frontier Biology, Graduate School of Dentistry, Osaka University, Suita 565-0871, Japan Received November 16, 2009; Accepted December 21, 2009 Abstract. Periodontal diseases, some of the most common infectious diseases seen in humans, are characterized by gingival inflammation, as well as loss of connective tissue and bone from around the roots of the teeth, which leads to eventual tooth exfoliation. In the past decade, the as- sociation of periodontal diseases with the development of systemic diseases has received increas- ing attention. Although a number of studies have presented evidence of close relationships between periodontal and systemic diseases, the majority of findings are limited to epidemiological studies, while the etiological details remain unclear. Nevertheless, a variety of recent hypothesis driven investigations have compiled various results showing that periodontal infection and subsequent direct oral-hematogenous spread of bacteria are implicated inPROOF the development of various systemic diseases. Herein, we present current understanding in regard to the relationship between periodon- tal and systemic diseases, including cardiovascular diseases, preterm delivery of low birth weight, diabetes mellitus, respiratory diseases, and osteoporosis. Keywords: periodontal disease, diabetes mellitus, cardiovascular disease, preterm delivery, Porphyromonas gingivalis, oral bacteria 1. Introduction detected in heart valve lesions and atheromatous plaque (5, 6), amniotic fluid of pregnant women with threatened Epidemiological and interventional studies of humans premature labor (11), and placentas from cases of preterm have revealed close associations between periodontal delivery (12, 13). -

The Mysterious Orphans of Mycoplasmataceae
The mysterious orphans of Mycoplasmataceae Tatiana V. Tatarinova1,2*, Inna Lysnyansky3, Yuri V. Nikolsky4,5,6, and Alexander Bolshoy7* 1 Children’s Hospital Los Angeles, Keck School of Medicine, University of Southern California, Los Angeles, 90027, California, USA 2 Spatial Science Institute, University of Southern California, Los Angeles, 90089, California, USA 3 Mycoplasma Unit, Division of Avian and Aquatic Diseases, Kimron Veterinary Institute, POB 12, Beit Dagan, 50250, Israel 4 School of Systems Biology, George Mason University, 10900 University Blvd, MSN 5B3, Manassas, VA 20110, USA 5 Biomedical Cluster, Skolkovo Foundation, 4 Lugovaya str., Skolkovo Innovation Centre, Mozhajskij region, Moscow, 143026, Russian Federation 6 Vavilov Institute of General Genetics, Moscow, Russian Federation 7 Department of Evolutionary and Environmental Biology and Institute of Evolution, University of Haifa, Israel 1,2 [email protected] 3 [email protected] 4-6 [email protected] 7 [email protected] 1 Abstract Background: The length of a protein sequence is largely determined by its function, i.e. each functional group is associated with an optimal size. However, comparative genomics revealed that proteins’ length may be affected by additional factors. In 2002 it was shown that in bacterium Escherichia coli and the archaeon Archaeoglobus fulgidus, protein sequences with no homologs are, on average, shorter than those with homologs [1]. Most experts now agree that the length distributions are distinctly different between protein sequences with and without homologs in bacterial and archaeal genomes. In this study, we examine this postulate by a comprehensive analysis of all annotated prokaryotic genomes and focusing on certain exceptions. -

MIB–MIP Is a Mycoplasma System That Captures and Cleaves Immunoglobulin G
MIB–MIP is a mycoplasma system that captures and cleaves immunoglobulin G Yonathan Arfia,b,1, Laetitia Minderc,d, Carmelo Di Primoe,f,g, Aline Le Royh,i,j, Christine Ebelh,i,j, Laurent Coquetk, Stephane Claveroll, Sanjay Vasheem, Joerg Joresn,o, Alain Blancharda,b, and Pascal Sirand-Pugneta,b aINRA (Institut National de la Recherche Agronomique), UMR 1332 Biologie du Fruit et Pathologie, F-33882 Villenave d’Ornon, France; bUniversity of Bordeaux, UMR 1332 Biologie du Fruit et Pathologie, F-33882 Villenave d’Ornon, France; cInstitut Européen de Chimie et Biologie, UMS 3033, University of Bordeaux, 33607 Pessac, France; dInstitut Bergonié, SIRIC BRIO, 33076 Bordeaux, France; eINSERM U1212, ARN Regulation Naturelle et Artificielle, 33607 Pessac, France; fCNRS UMR 5320, ARN Regulation Naturelle et Artificielle, 33607 Pessac, France; gInstitut Européen de Chimie et Biologie, University of Bordeaux, 33607 Pessac, France; hInstitut de Biologie Structurale, University of Grenoble Alpes, F-38044 Grenoble, France; iCNRS, Institut de Biologie Structurale, F-38044 Grenoble, France; jCEA, Institut de Biologie Structurale, F-38044 Grenoble, France; kCNRS UMR 6270, Plateforme PISSARO, Institute for Research and Innovation in Biomedicine - Normandie Rouen, Normandie Université, F-76821 Mont-Saint-Aignan, France; lProteome Platform, Functional Genomic Center of Bordeaux, University of Bordeaux, F-33076 Bordeaux Cedex, France; mJ. Craig Venter Institute, Rockville, MD 20850; nInternational Livestock Research Institute, 00100 Nairobi, Kenya; and oInstitute of Veterinary Bacteriology, University of Bern, CH-3001 Bern, Switzerland Edited by Roy Curtiss III, University of Florida, Gainesville, FL, and approved March 30, 2016 (received for review January 12, 2016) Mycoplasmas are “minimal” bacteria able to infect humans, wildlife, introduced into naive herds (8). -

Genomic Islands in Mycoplasmas
G C A T T A C G G C A T genes Review Genomic Islands in Mycoplasmas Christine Citti * , Eric Baranowski * , Emilie Dordet-Frisoni, Marion Faucher and Laurent-Xavier Nouvel Interactions Hôtes-Agents Pathogènes (IHAP), Université de Toulouse, INRAE, ENVT, 31300 Toulouse, France; [email protected] (E.D.-F.); [email protected] (M.F.); [email protected] (L.-X.N.) * Correspondence: [email protected] (C.C.); [email protected] (E.B.) Received: 30 June 2020; Accepted: 20 July 2020; Published: 22 July 2020 Abstract: Bacteria of the Mycoplasma genus are characterized by the lack of a cell-wall, the use of UGA as tryptophan codon instead of a universal stop, and their simplified metabolic pathways. Most of these features are due to the small-size and limited-content of their genomes (580–1840 Kbp; 482–2050 CDS). Yet, the Mycoplasma genus encompasses over 200 species living in close contact with a wide range of animal hosts and man. These include pathogens, pathobionts, or commensals that have retained the full capacity to synthesize DNA, RNA, and all proteins required to sustain a parasitic life-style, with most being able to grow under laboratory conditions without host cells. Over the last 10 years, comparative genome analyses of multiple species and strains unveiled some of the dynamics of mycoplasma genomes. This review summarizes our current knowledge of genomic islands (GIs) found in mycoplasmas, with a focus on pathogenicity islands, integrative and conjugative elements (ICEs), and prophages. Here, we discuss how GIs contribute to the dynamics of mycoplasma genomes and how they participate in the evolution of these minimal organisms. -

Serological Evidence That Chlamydiae and Mycoplasmas Are Involved in Infertility of Women B
Serological evidence that chlamydiae and mycoplasmas are involved in infertility of women B. R. M\l=o/\ller,D. Taylor-Robinson, Patricia M. Furr, B. Toft and J. Allen Division of Sexually Transmitted Diseases, MRC Clinical Research Centre, Watford Road, Harrow, Middlesex HAI 3UJ, U.K., and ^Department of Obstetrics and Gynaecology, University of Aarhus, DK-8000, Aarhus, Denmark Summary. Women with a history of infertility for 2 or more years were examined by hysterosalpingography (HSG) and antibodies against Chlamydia trachomatis, Myco- plasma hominis and M. genitalium were measured by a microimmunofluorescence technique in sera obtained immediately before HSG. Of 45 women with abnormal HSG findings, 15 (33%) had antibodies to C. trachomatis and 16 (35\m=.\5%)to M. hominis. In contrast, of 61 women with normal HSG findings, only 8 (13%) and 7 (11\m=.\5%)had antibodies to these micro-organisms, respectively. Antibody against M. genitalium was found in 26 of the patients (20% abnormal HSG and 28% normal HSG), indicating the need for further investigation of the significance of this mycoplasma in female infertility. The present results do confirm, however, that C. trachomatis is an important cause of infertility in women and suggest strongly that M. hominis is implicated. Introduction Infertility in women is caused often by tubai damage after pelvic inflammatory disease. Chlamydia trachomatis is a well-known pathogen in upper genital-tract infections and accounts for 25-50% of all cases of pelvic inflammatory disease (Paavonen, 1979) while Mycoplasma hominis is believed to be responsible for about 25% of all the cases (Moller, 1983). -

Fis-His-2020-Abstract-Supplement-Free-Paper.Pdf
Abstract supplement (free paper abstracts) FIS/HIS International 2020 cannot be held responsible for errors or inconsistencies contained within the abstract supplement. Only major formatting alterations have been made and abstract content remains consistent with what was entered at time of submission by the author/s. How to search this document Laptop, PC or similar: ‘Ctrl’ + ‘F’ or ‘Edit’ then ‘Find’ Smart device: Open this PDF in either the 'iBooks' app (Apple) or 'My Files' app (Android). Both are built in apps and have a magnifying glass search icon. Contents A quick reference guide by topic, paper number and presentation type can be found at the end of the document or click here. (Click paper title to take you straight there) Free paper oral presentations 11: Investigations, actions and learning from an outbreak of SARS-CoV-2 infection among health care workers in the United Kingdom ........................................................................................................................ 13 12: Procalcitonin as an antibiotic stewardship tool in COVID-19 patients in the intensive care ...................... 14 20: Impact of antibiotic timing on mortality from Gram negative Bacteraemia in an English District General Hospital: the importance of getting it right every time .................................................................................... 15 35: Case report: A fall in the forest ................................................................................................................... 16 65: Not -

Correlation Between the Oral Microbiome and Brain Resting State Connectivity in Smokers
bioRxiv preprint doi: https://doi.org/10.1101/444612; this version posted October 16, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Correlation between the oral microbiome and brain resting state connectivity in smokers Dongdong Lin1, Kent Hutchison2, Salvador Portillo3, Victor Vegara1, Jarod Ellingson2, Jingyu Liu1,3, Amanda Carroll-Portillo3,* ,Vince D. Calhoun1,3,* 1The Mind Research Network, Albuquerque, New Mexico, 87106 2University of Colorado Boulder, Boulder, CO 3University of New Mexico, Department of Electrical and Computer Engineering, Albuquerque, New Mexico, 87106 * authors contributed equally to the work. Abstract Recent studies have shown a critical role for the gastrointestinal microbiome in brain and behavior via a complex gut–microbiome–brain axis, however, the influence of the oral microbiome in neurological processes is much less studied, especially in response to the stimuli in the oral microenvironment such as smoking. Additionally, given the complex structural and functional networks in brain system, our knowledge about the relationship between microbiome and brain functions on specific brain circuits is still very limited. In this pilot work, we leverage next generation microbial sequencing with functional MRI techniques to enable the delineation of microbiome-brain network links as well as their relations to cigarette smoking. Thirty smokers and 30 age- and sex- matched non-smokers were recruited for measuring both microbial community and brain functional networks. Statistical analyses were performed to demonstrate the influence of smoking on: the taxonomy and abundance of the constituents within the oral microbial community, brain functional network connectivity, and associations between microbial shifts and the brain signaling network. -

CASEE-2017 Session1b -Eva-Tvrda
A The 8th International CASEE Conference Warsaw University of Life Sciences – SGGW May 14 - 16, 2017 . Decreased sperm quality visible in routine semen analysis: . loss of sperm motility . morphological alterations . acrosome dysfunction . disruption of membrane integrity . oxidative stress . Most data connected to bacterial contamination of ejaculates: well-known causative agents of urogenital tract infections . Escherichia coli, Staphylococcus aureus, Ureaplasma urealyticum, Mycoplasma hominis, Chlamydia trachomatis . Ejaculates collected for reproductive technologies - certain contamination: . semen collection is not an entirely serile process . factors for semen contamination: artifical vaginas, environmental conditions, human factors . Current interest shifts to other bacteria, responsible for the colonization and contamination of the male urogenital tract, rather than infection . Gram-positive, catalase-negative, non-spore-forming, facultative anaerobic bacteria . Lactic acid bacteria (LAB) that produce bacteriocins . Origins: environmental, animal and human sources . E. faecalis: . most common in the gastrointestinal tract, and may be found in human and animal faeces . associated with clinical urinary tract infections, hepatobiliary sepsis, endocarditis, surgical wound infection, bacteraemia and neonatal sepsis . able to survive a range of adverse environments allowing multiple routes of cross- contamination . resistant to a broad range of antibiotics including ampicillin, ciprofloxacin and imipenem ANTIBIOTICS NATURALLY OCCURING -

GBS 10 Strep11 Other 60 Vaginal Swabs! Real-Time LAMP!
• Specificity studies were performed using a panel of 10 Group B Streptococcus agalactiae strains, 11 of the most commonly occurring Streptococcus genus strains and 60 other organisms commonly found in the human genital tract. • One hundred and twenty four (124) vaginal swabs sampled from pregnant women were tested with the real-time LAMP assay and compared to a real- time PCR assay (5). Neonatal infections are most commonly caused by Group B Streptococcus GBS 10 (Streptococcus agalactiae) (1). These infections can be classified as either early on-set • Of the 124 vaginal swabs, 85 tested positive for Group B Streptococcus and & or late-onset disease. Early onset infection (EOI) occurs during the first week of life 39 tested negative. with late onset infection (LOI) occurring between one week and three months. It is Strep11 thought that early onset disease develops in the foetus after aspiration of infected • Comparable results were achieved by both the real-time PCR and real –time amniotic fluid (2). Late onset infection is less well understood, and while passage LAMP assays. Results are listed in Table 4. Also shown in the table are through the birth canal is an obvious route of infection, it is also thought that community previously obtained microbiological culture data for the isolation and sources are involved (2). Other identification of GBS. 60 One approach used to identify potential for infection is a screening of pregnant women, where prophylaxis is offered to those identified as carriers (3). This approach is also • Figure 1: Breakdown of specificity panels investigated during the development of the Table 2: Results of GBS testing of vaginal swabs. -

Research Article Rapid PCR Detection of Mycoplasma Hominis, Ureaplasma Urealyticum,Andureaplasma Parvum
Hindawi Publishing Corporation International Journal of Bacteriology Volume 2013, Article ID 168742, 7 pages http://dx.doi.org/10.1155/2013/168742 Research Article Rapid PCR Detection of Mycoplasma hominis, Ureaplasma urealyticum,andUreaplasma parvum Scott A. Cunningham,1 Jayawant N. Mandrekar,2 Jon E. Rosenblatt,1 and Robin Patel1,3 1 Division of Clinical Microbiology, Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, MN 55905, USA 2 Division of Biomedical Statistics and Informatics, Department of Health Science Research, Mayo Clinic, Rochester, MN 55905, USA 3 Division of Infectious Diseases, Department of Medicine, Mayo Clinic, Rochester, MN 55905, USA Correspondence should be addressed to Robin Patel; [email protected] Received 5 November 2012; Accepted 30 January 2013 Academic Editor: Sam R. Telford Copyright © 2013 Scott A. Cunningham et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Objective. We compared laboratory developed real-time PCR assays for detection of Mycoplasma hominis and for detection and differentiation of Ureaplasma urealyticum and parvum to culture using genitourinary specimens submitted for M. hominis and Ureaplasma culture. Methods. 283 genitourinary specimens received in the clinical bacteriology laboratory for M. hominis and Ureaplasma species culture were evaluated. Nucleic acids were extracted using the Total Nucleic Acid Kit on the MagNA Pure 2.0. 5 L of the extracts were combined with 15 L of each of the two master mixes. Assays were performed on the LightCycler 480 II system. Culture was performed using routine methods. -
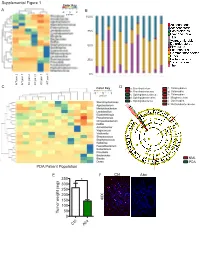
Mental Figure 1 Color Key a -2 0 2 B Z-Score 100%
Supplemental Figure 1 Color Key A -2 0 2 B z-score 100% 75% 50% 25% 0% KC pan 1 WT pan 3 WT KC pan 3 WT pan 2 WT pan 1 WT KC pan 2 C Color Key D a: Brevibacterium f: Chlamydiales b: Brevibacteriaceae g: Chlamydiia -3 0 3 z-score c: Sphingobacteriaceae h: Chlamydiae d: Sphingobacteriales i: Mogibacterium e: Sphingobacteriia j: Oscillospira k: Methylobacteriaceae NML Control Microb.-entrained MΦ PDA PDA Patient Population Control Microb.-entrained MΦ + Myd88i E F Ctrl Abx 350 * 300 250 200 150 40X 100 Tumor weight (mg) 50 0 x Ctrl Ab Supplemental Figure 2 A KC WT B ** * Actinobacteria * ** Bacteroidetes Cyanobacteria Deferribacteres * Firmicutes Proteobacteria % Relative abundance TM7 Others Time(wks) 3 9 13 16 20 24 28 32 36 3 9 13 16 20 24 28 32 36 Alpha Diversity Measure C E 60 KC WT 40 20 B. pseudolongum B. animalis 60 5 KC WT 0 B. adolescentis 40% Rel. abundance 3 9 13 16 20 24 28 32 36 3 9 13 16 20 24 28 32 36 20 Age (weeks) B. pseudolongum B. animalis 5 0 B. adolescentis % Rel. abundance 3 9 13 16 20 24 28 32 36 3 9 13 16 20 24 28 32 36 F Age (weeks) Week 3 Week 9 Week 13 p=0.678 p=0.02 p=0.385 Time(wks) 3 9 24 20 13 16 D 28 32 36 Week 13 KC WT Firmicutes; Ruminococcus Firmicutes; Dehalobacterium Alpha Diversity Measure Firmicutes; Oscillospira Bacteroidates; Odoribacter Axis.2 [12.7%] Actinobacteria; Bifidobacterium Axis.2 [23.8%] Axis.2 [24.7%] Week 16 Bacteroidetes; Bacteroidales Axis.1 [80.8%] Axis.1 [65.4%] Axis.1 [49.6%] Actinobacteria; Bifidobacterium Week 16 Week 20 Week 24 Week 20 p=0.339 p=0.036 p=0.021 Firmicutes; Dehalobacterium