BD™ Gardnerella Selective Agar with 5% Human Blood
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Human Microbiota Network: Unveiling Potential Crosstalk Between the Different Microbiota Ecosystems and Their Role in Health and Disease
nutrients Review Human Microbiota Network: Unveiling Potential Crosstalk between the Different Microbiota Ecosystems and Their Role in Health and Disease Jose E. Martínez †, Augusto Vargas † , Tania Pérez-Sánchez , Ignacio J. Encío , Miriam Cabello-Olmo * and Miguel Barajas * Biochemistry Area, Department of Health Science, Public University of Navarre, 31008 Pamplona, Spain; [email protected] (J.E.M.); [email protected] (A.V.); [email protected] (T.P.-S.); [email protected] (I.J.E.) * Correspondence: [email protected] (M.C.-O.); [email protected] (M.B.) † These authors contributed equally to this work. Abstract: The human body is host to a large number of microorganisms which conform the human microbiota, that is known to play an important role in health and disease. Although most of the microorganisms that coexist with us are located in the gut, microbial cells present in other locations (like skin, respiratory tract, genitourinary tract, and the vaginal zone in women) also play a significant role regulating host health. The fact that there are different kinds of microbiota in different body areas does not mean they are independent. It is plausible that connection exist, and different studies have shown that the microbiota present in different zones of the human body has the capability of communicating through secondary metabolites. In this sense, dysbiosis in one body compartment Citation: Martínez, J.E.; Vargas, A.; may negatively affect distal areas and contribute to the development of diseases. Accordingly, it Pérez-Sánchez, T.; Encío, I.J.; could be hypothesized that the whole set of microbial cells that inhabit the human body form a Cabello-Olmo, M.; Barajas, M. -
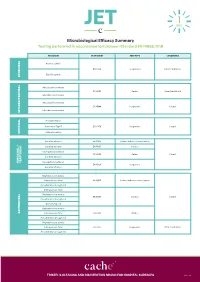
JET Microbiological Efficacy Summary
Microbiological Efficacy Summary Testing performed in accordance to European Standard EN 14885:2018 ORGANISM TEST NORM TEST TYPE CONDITIONS Bacillus subtilis EN 13704 Suspension Clean 1 and Dirty 1 Bacillus cereus SPORICIDAL Mycobacterium terrae EN 14563 Carrier Clean 1 and Dirty 2 Mycobacterium avium Mycobacterium terrae EN 14348 Suspension Clean 1 Mycobacterium avium MYCOBACTERICIDAL Poliovirus Type 1 Adenovirus Type 5 EN 14476 Suspension Clean 1 Murine Norovirus VIRUCIDAL Candida albicans EN 16615 Surface with mechanical action Candida albicans EN 13697 Surface Aspergillus brasiliensis EN 14562 Carrier Clean 1 Candida albicans YEASTICIDAL FUNGICIDAL / FUNGICIDAL Aspergillus brasiliensis EN 13624 Suspension Candida albicans Staphylococcus aureus Enterococcus hirae EN 16615 Surface with mechanical action Pseudomonas aeruginosa Enterococcus hirae Staphylococcus aureus EN 13697 Surface Clean 1 Pseudomonas aeruginosa Escherichia coli Staphylococcus aureus BACTERICIDAL Enterococcus hirae EN 14561 Carrier Pseudomonas aeruginosa Staphylococcus aureus Enterococcus hirae EN 13727 Suspension Clean 1 and Dirty 1 Pseudomonas aeruginosa TRISTEL’S CLEANING AND DISINFECTION BRAND FOR HOSPITAL SURFACES Page 1 of 3 Additional Testing TEST METHOD RNA DNA / Polyacrylamide gel electrophoresis (PAGE) ORGANISM TEST METHOD TEST TYPE CONDITIONS Acanthamoeba castellanii cysts Following the method of EN 13704 Suspension Clean 1 PROTOZOA Bacillus subtilis EN 17126 Suspension Clean 1 Bacillus cereus Clostridium difficile EN 13704 Suspension Clean 1 and Dirty 1 -

NOTES in Vitro Activities of Norfloxacin and Ciprofloxacin Against
ANTIMICROBIAL AGENTS AND CHEMOTHERAPY, July 1984, p. 94-96 Vol. 26, No. 1 0066-4804/84/070094-03$02.00/0 Copyright C 1984, American Society for Microbiology NOTES In Vitro Activities of Norfloxacin and Ciprofloxacin Against Mycobacterium tuberculosis, M. avium Complex, M. chelonei, M. fortuitum, and M. kansasii J. DOUGLAS GAY, DONALD R. DEYOUNG, AND GLENN D. ROBERTS* Section of Clinical Microbiology, Department of Laboratory Medicine, Mayo Clinic and Mayo Foundation, Rochester, Minnesota 55905 Received 28 November 1983/Accepted 4 April 1984 The activities of ciprofloxacin and norfloxacin against 100 mycobacteria isolates were studied in vitro by the 1% standard proportion method. Ciprofloxacin was more active against M. tuberculosis and M. fortuitum with MICs of 1.0 and 0.25 ,ug/ml, respectively, against 90% of isolates; norfloxacin had MICs of 8.0 and 2.0 ,ug/ml, respectively, against 90% of isolates. Nalidixic acid and other heterocyclic carbonic acid deriva- studied. The organisms were taken from the Mayo Clinic tives have been used primarily in the treatment of urinary stock culture collection, which included recent clinical iso- tract infections for many years. The compounds of this lates. Stock cultures were maintained on Middlebrook 7H10 general group include nalidixic acid, oxolinic acid, pipemidic agar slants (Difco Laboratories, Detroit, Mich.) and were acid, cinoxacin, and rosoxacin. Two new substances in this subcultured monthly. The identification of isolates was series which have been recently synthesized are norfloxacin based on standard biochemical tests (17) and gas-liquid (6) (1-ethyl-6-fluoro-1,4-dihydro-4-oxo-7-[ 1-piperazinyl ]-3- chromatography (16). -

A Genomic Journey Through a Genus of Large DNA Viruses
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Virology Papers Virology, Nebraska Center for 2013 Towards defining the chloroviruses: a genomic journey through a genus of large DNA viruses Adrien Jeanniard Aix-Marseille Université David D. Dunigan University of Nebraska-Lincoln, [email protected] James Gurnon University of Nebraska-Lincoln, [email protected] Irina V. Agarkova University of Nebraska-Lincoln, [email protected] Ming Kang University of Nebraska-Lincoln, [email protected] See next page for additional authors Follow this and additional works at: https://digitalcommons.unl.edu/virologypub Part of the Biological Phenomena, Cell Phenomena, and Immunity Commons, Cell and Developmental Biology Commons, Genetics and Genomics Commons, Infectious Disease Commons, Medical Immunology Commons, Medical Pathology Commons, and the Virology Commons Jeanniard, Adrien; Dunigan, David D.; Gurnon, James; Agarkova, Irina V.; Kang, Ming; Vitek, Jason; Duncan, Garry; McClung, O William; Larsen, Megan; Claverie, Jean-Michel; Van Etten, James L.; and Blanc, Guillaume, "Towards defining the chloroviruses: a genomic journey through a genus of large DNA viruses" (2013). Virology Papers. 245. https://digitalcommons.unl.edu/virologypub/245 This Article is brought to you for free and open access by the Virology, Nebraska Center for at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Virology Papers by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Authors Adrien Jeanniard, David D. Dunigan, James Gurnon, Irina V. Agarkova, Ming Kang, Jason Vitek, Garry Duncan, O William McClung, Megan Larsen, Jean-Michel Claverie, James L. Van Etten, and Guillaume Blanc This article is available at DigitalCommons@University of Nebraska - Lincoln: https://digitalcommons.unl.edu/ virologypub/245 Jeanniard, Dunigan, Gurnon, Agarkova, Kang, Vitek, Duncan, McClung, Larsen, Claverie, Van Etten & Blanc in BMC Genomics (2013) 14. -

Sample Report Vaginosis.Pdf
LAB #: Sample Report CLIENT #: 12345 PATIENT: Sample Patient DOCTOR: Sample Doctor ID: Doctor's Data, Inc. SEX: Female 3755 Illinois Ave. DOB: 01/01/1993 AGE: 25 St. Charles, IL 60174 U.S.A. !"#$%&'(%(!)*'+%,- GRAM STAIN MICROSCOPY BACTERIAL VAGINOSIS SCORE Normal Abnormal Expected score interpretation: Lactobacilli None Mod - Many 0 - 3 BV not likely 4 - 6 BV indeterminate Curved Gram 10 7-10 BV highly suggestive Negative Rods Many None Small Gram 1 Many None The BV score is calculated based upon the gram stain results Negative Rods and is independent of the yeast, and bacterial cultures. 1Nugent Scoring System. (Nugent et al. J. Clin. Micro. Yeast None None (1991)29:297-301) RBC’s None None YEAST CULTURE 2+ Candida albicans WBC’s 0 0 - 6 Clue Cells Present None Eosinophils N/A None Eosinophils reported and Wrights Stain performed when WBC’s >6 Additional Gram Stain Findings: Rare Gram positive cocci in pairs BACTERIOLOGY CULTURE Expected/Beneficial flora Commensal (Imbalanced) flora Dysbiotic flora NG Lactobacillus spp. 2+ Alpha hemolytic strep 3+ Gardnerella vaginalis 2+ Gamma hemolytic strep 4+ Enterococcus faecalis 1+ Staphylococcus not aureus NG = No Growth SPECIMEN DATA Comments: Date Collected: 01/14/2019 Date Received: 01/16/2019 Date Completed: 01/23/2019 ©Doctor’s Data, Inc. !!! ADDRESS: 3755 Illinois Avenue, St. Charles, IL 60174-2420 !!! MED DIR: Erlo Roth, MD !!! CLIA ID NO: 14D0646470 0002038 LAB #: Sample Report CLIENT #: 12345 PATIENT: Sample DOCTOR: Sample Doctor Patient ID: Doctor's Data, Inc. SEX: Female 3755 Illinois Ave. DOB: 01/01/1993 St. Charles, IL 60174 U.S.A. -

Mycobacterium Avium Possesses Extracellular DNA That Contributes to Biofilm Formation, Structural Integrity, and Tolerance to Antibiotics
Mycobacterium avium Possesses Extracellular DNA that Contributes to Biofilm Formation, Structural Integrity, and Tolerance to Antibiotics Rose, S. J., Babrak, L. M., & Bermudez, L. E. (2015). Mycobacterium avium Possesses Extracellular DNA that Contributes to Biofilm Formation, Structural Integrity, and Tolerance to Antibiotics. PLoS ONE, 10(5), e0128772. doi: 10.1371/journal.pone.0128772 10.1371/journal.pone.0128772 Public Library of Science Version of Record http://cdss.library.oregonstate.edu/sa-termsofuse RESEARCH ARTICLE Mycobacterium avium Possesses Extracellular DNA that Contributes to Biofilm Formation, Structural Integrity, and Tolerance to Antibiotics Sasha J. Rose1,2, Lmar M. Babrak1,2, Luiz E. Bermudez1,2* 1 Department of Biomedical Sciences, College of Veterinary Medicine, Oregon State University, Corvallis, Oregon, United States of America, 2 Department of Microbiology, College of Science, Oregon State University, Corvallis, Oregon, United States of America * [email protected] Abstract Mycobacterium avium subsp. hominissuis is an opportunistic pathogen that is associated with biofilm-related infections of the respiratory tract and is difficult to treat. In recent years, extracellular DNA (eDNA) has been found to be a major component of bacterial biofilms, in- OPEN ACCESS cluding many pathogens involved in biofilm-associated infections. To date, eDNA has not Citation: Rose SJ, Babrak LM, Bermudez LE (2015) been described as a component of mycobacterial biofilms. In this study, we identified and Mycobacterium avium Possesses Extracellular DNA characterized eDNA in a high biofilm-producing strain of Mycobacterium avium subsp. that Contributes to Biofilm Formation, Structural hominissuis (MAH). In addition, we surveyed for presence of eDNA in various MAH strains Integrity, and Tolerance to Antibiotics. -

Characterization of an Α-Glucosidase Enzyme Conserved in Gardnerella
bioRxiv preprint doi: https://doi.org/10.1101/2020.05.11.086124; this version posted May 11, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Characterization of an a-glucosidase enzyme conserved in Gardnerella spp. isolated 2 from the human vaginal microbiome 3 4 Pashupati Bhandari1, Jeffrey P. Tingley2, D. Wade Abbott2 and Janet E. Hill1,* 5 6 1Department of Veterinary Microbiology, Western College of Veterinary Medicine, 7 University of Saskatchewan, 52 Campus Drive, Saskatoon, Saskatchewan, S7N 5B4, 8 Canada 9 2Lethbridge Research and Development Centre, Agriculture and Agri-Food Canada, 10 Lethbridge, Alberta, T1J 4B1, Canada 11 12 *To whom correspondence should be addressed 13 [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.05.11.086124; this version posted May 11, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 14 Abstract 15 Gardnerella spp. in the vaginal microbiome are associated with bacterial vaginosis, a 16 dysbiosis in which lactobacilli dominant microbial community is replaced with mixed 17 aerobic and anaerobic bacteria including Gardnerella species. The co-occurrence of 18 multiple Gardnerella species in the vaginal environment is common, but different species 19 are dominant in different women. -

Gardnerella Vaginalis: Characteristics, Clinical Considerations, and Controversies B
CLINICAL MICROBIOLOGY REVIEWS, July 1992, p. 213-237 Vol. 5, No. 3 0893-8512/92/030213-25$02.00/0 Copyright 1992, American Society for Microbiology Gardnerella vaginalis: Characteristics, Clinical Considerations, and Controversies B. WESLEY CATLINt Department ofMicrobiology, The Medical College of Wisconsin, Milwaukee, Wisconsin 53226 INTRODUCTION .................................................................. .214 IDENTIFICATION AND CHARACTERISTICS OF G. VAGINALIS ....... 0........... *** ...... 00 .............. *..214 Appearance of Cells and Colonies........................................... ........ 000 ........ 000 ........... 0.0 .........0.214 1%.9 1- Isolation Methods................................................................ ............... oo ........................ *..* ...215 Differential Tests................................................................. ..216 It . Presumptive identification.................................................. I.............................. .. 1lo ^.,1IA Confirmation................%.,%FARRAR 21143IMPRA . o . 0 o 0 . o . o o o o 0 o........... ............. 00.00 ....................................................__________ _ _ klosn Confusing catalase-negative bacteria........ .216 Methods: some yield inconsistent results... .217 1% s _ Other characteristics............................ ......... ......o. ... .2177 Rapid detection................................... .2:,17 Structure, Composition, and Toxic Products ;... ****O..* ....... 0.000.0 .................*.................................z'17 -

Examining the Link Between Macrophyte Diversity, Bacterial
EXAMINING THE LINK BETWEEN MACROPHYTE DIVERSITY, BACTERIAL DIVERSITY, AND DENITRIFICATION FUNCTION IN WETLANDS DISSERTATION Presented in Partial Fulfillment of the Requirements for The Degree of Doctor of Philosophy in the Graduate School of The Ohio State University By Janice M. Gilbert, B.E.S., B.Ed., M.E.S., M.S. ***** The Ohio State University 2004 Dissertation Committee: Professor Virginie Bouchard, Adviser Approved by Professor Serita D. Frey, Co-adviser Professor Olli H. Tuovinen Professor Frederick C. Michel, Jr. Adviser Environmental Science Graduate Program ABSTRACT The relationship between aquatic plant (macrophyte) diversity, bacterial diversity, and the biochemical reduction of nitrate (denitrification) within wetlands was examined. Denitrification occurs under anoxic conditions when nitrate is reduced to either nitrous oxide (N2O), or dinitrogen (N2). Although previous studies have identified physical and chemical factors regulating the production of either gas in wetlands, the role that macrophyte diversity plays in this process is not known. The central hypothesis, based on the niche-complimentarity mechanism, was that an increase in macrophyte diversity would lead to increased bacterial diversity, increased denitrification, and decreased N2O flux. This hypothesis was investigated in two mesocosm studies to control environmental conditions while altering macrophyte functional groups (FG) and functional group diversity. In Study #1, five macrophyte functional groups (clonal dominants, tussocks, reeds, facultative annuals, and obligate annuals) were each represented by two species. Fifty-five mesocosms with 5-6 replicates of 0, 1, 2, 3, 4, or 5 macrophyte FG (0-10 species) were established in the spring of 2001 and sampled in August 2001, September 2001, and April 2002. -

The Evolution of a Gene Cluster Containing a Plant-Like Protein In
EVOLUTION OF A PLANT-LIKE GENE ANCIENTLY ACQUIRED AS PART OF A GENOMIC ISLAND IN XANTHOMONAS A DISSERTATION SUBMITTED TO THE GRADUATE DIVISION OF THE UNIVERSITY OF HAWAI‘I AT MᾹNOA IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY IN MOLECULAR BIOSCIENCES AND BIOENGINEERING May 2012 BY KEVIN SCHNEIDER DISSERTATION COMMITTEE GERNOT PRESTING, CHAIRPERSON ANNE ALVAREZ YANGRAE CHO GUYLAINE POISSON SEAN CALLAHAN Dedicated to my Parents! i Acknowledgments I want to give my biggest thanks to Dr Gernot Presting for providing me with so many opportunities during my career at UH Manoa. The teaching assistantship I received on an unexpected short notice that began my PhD to working and publishing on exciting and interesting topics from corn centromeres to bacterial genomes. I am forever grateful for the time, patience, and energy he has spent mentoring me. This work would not have been possible without Dr Anne Alvarez. She has provided not only her knowledge of plant pathology, but also her collection of bacterial strains that the majority of my research required. Also, I thank Asoka Da Silva whom has provided his expertise and skills to culture and purify the hundreds of strains used in this study. The analysis in this work would not have begun without the initial phylogenomic analysis of Arabidopsis completed by Aren Ewing. His work laid the foundation to stick with studying bacterial genomic evolution in light of all of the wonderful work to study the genomic evolution of the centromeres of Zea mays in our lab. I also thank all of my lab mates Anupma Sharma, Thomas Wolfgruber, Jamie Allison, Jeffrey Lai, Megan Nakashima, Ronghui Xu, Zidian Xie, Grace Kwan, Margaret Ruzicka, Krystle Salazar and Erin Mitsunaga from the past and the present for their advice, help, discussions and their friendship and casual chit-chat. -

Microbiome Profile of the Amniotic Fluid As a Predictive Biomarker Of
www.nature.com/scientificreports OPEN Microbiome profle of the amniotic fuid as a predictive biomarker of perinatal outcome Received: 11 May 2017 Daichi Urushiyama1,2, Wataru Suda3,4, Eriko Ohnishi1, Ryota Araki2, Chihiro Kiyoshima2, Accepted: 29 August 2017 Masamitsu Kurakazu2, Ayako Sanui5, Fusanori Yotsumoto2, Masaharu Murata5, Kazuki Published: xx xx xxxx Nabeshima6, Shin’ichiro Yasunaga7, Shigeru Saito8, Makoto Nomiyama9, Masahira Hattori3,10, Shingo Miyamoto2 & Kenichiro Hata1 Chorioamnionitis (CAM), an infammation of the foetal membranes due to infection, is associated with preterm birth and poor perinatal prognosis. The present study aimed to determine whether CAM can be diagnosed prior to delivery based on the bacterial composition of the amniotic fuid (AF). AF samples from 79 patients were classifed according to placental infammation: Stage III (n = 32), CAM; Stage II (n = 27), chorionitis; Stage 0-I (n = 20), sub-chorionitis or no neutrophil infltration; and normal AF in early pregnancy (n = 18). Absolute quantifcation and sequencing of 16S rDNA showed that in Stage III, the 16S rDNA copy number was signifcantly higher and the α-diversity index lower than those in the other groups. In principal coordinate analysis, Stage III formed a separate cluster from Stage 0-I, normal AF, and blank. Forty samples were classifed as positive for microbiomic CAM (miCAM) defned by the presence of 11 bacterial species that were found to be signifcantly associated with CAM and some parameters of perinatal prognosis. The diagnostic accuracy for CAM according to miCAM was: sensitivity, approximately 94%, and specifcity, 79–87%. Our fndings indicate the possibility of predicting CAM prior to delivery based on the AF microbiome profle. -

A Cluster Analysis of Bacterial Vaginosis–Associated Microflora
American Journal of Epidemiology Vol. 162, No. 6 Copyright ª 2005 by the Johns Hopkins Bloomberg School of Public Health Printed in U.S.A. All rights reserved DOI: 10.1093/aje/kwi243 A Cluster Analysis of Bacterial Vaginosis–associated Microflora and Pelvic Inflammatory Disease Roberta B. Ness1,2, Kevin E. Kip1, Sharon L. Hillier1,2, David E. Soper3, Carol A. Stamm4, Richard L. Sweet5, Peter Rice6, and Holly E. Richter7 Downloaded from https://academic.oup.com/aje/article/162/6/585/100561 by guest on 27 September 2021 1 Department of Epidemiology, Graduate School of Public Health, University of Pittsburgh, Pittsburgh, PA. 2 Department of Obstetrics, Gynecology, and Reproductive Sciences, Magee-Womens Hospital and Magee-Womens Research Institute, Pittsburgh, PA. 3 Department of Obstetrics and Gynecology, Medical University of South Carolina, Charleston, SC. 4 Department of Obstetrics and Gynecology, Denver Health Medical Center, Denver, CO. 5 Center for Women’s Health, University of California at Davis, San Diego, CA. 6 Division of Infectious Disease, Boston Medical Center, Maxwell Finland Laboratory, Boston, MA. 7 Department of Obstetrics and Gynecology, University of Alabama School of Medicine, Birmingham, AL. Received for publication January 7, 2005; accepted for publication April 28, 2005. Controversy surrounds the association between bacterial vaginosis (BV) and pelvic inflammatory disease (PID). Women (N ¼ 1,140) were ascertained at five US centers, enrolled (1999–2001), and followed up for a median of 3 years. Serial vaginal swabs were obtained for Gram’s stain and cultures. PID was defined as 1) histologic endometritis or 2) pelvic pain and tenderness plus oral temperature >38.8°C, leukorrhea or mucopus, erythrocyte sedimentation rate >15 mm/hour, white blood cell count >10,000, or gonococcal/chlamydial lower genital infection.