NOTES in Vitro Activities of Norfloxacin and Ciprofloxacin Against
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

BD™ Gardnerella Selective Agar with 5% Human Blood
INSTRUCTIONS FOR USE – READY-TO-USE PLATED MEDIA PA-254094.06 Rev.: July 2014 BD Gardnerella Selective Agar with 5% Human Blood INTENDED USE BD Gardnerella Selective Agar with 5% Human Blood is a partially selective and differential medium for the isolation of Gardnerella vaginalis from clinical specimens. PRINCIPLES AND EXPLANATION OF THE PROCEDURE Microbiological method. Gardnerella vaginalis is considered to be one of the organisms causing vaginitis.1-4 Although the organism may be present in a high percentage of normal women in the vaginal flora, its importance as a cause of non-specific vaginitis (also called bacterial vaginosis) has never been questioned. In symptomatic women, G. vaginalis frequently is associated with anaerobes such as Prevotella bivia, P. disiens, Mobiluncus, Peptostreptococcus, and/or others which are a regular part of the urethral or intestinal, but not vaginal flora. In non-specific vaginitis, normal Lactobacillus flora is reduced or absent. Gardnerella vaginalis is considered to be the indicator organism for non-specific vaginitis which, in fact, is a polymicrobial infection.3,4 Although non- culture methods such as a direct Gram stain have been recommended in recent years for genital specimens, culture is still preferred by many laboratories.1,5 G. vaginalis may also be responsible for a variety of other diseases such as preterm birth, chorioamnionitis, urinary tract infections, newborn infections, and septicemia.6 The detection of the organism on routinely used media is difficult since Gardnerella and other -

Computational Antibiotics Book
Andrew V DeLong, Jared C Harris, Brittany S Larcart, Chandler B Massey, Chelsie D Northcutt, Somuayiro N Nwokike, Oscar A Otieno, Harsh M Patel, Mehulkumar P Patel, Pratik Pravin Patel, Eugene I Rowell, Brandon M Rush, Marc-Edwin G Saint-Louis, Amy M Vardeman, Felicia N Woods, Giso Abadi, Thomas J. Manning Computational Antibiotics Valdosta State University is located in South Georgia. Computational Antibiotics Index • Computational Details and Website Access (p. 8) • Acknowledgements (p. 9) • Dedications (p. 11) • Antibiotic Historical Introduction (p. 13) Introduction to Antibiotic groups • Penicillin’s (p. 21) • Carbapenems (p. 22) • Oxazolidines (p. 23) • Rifamycin (p. 24) • Lincosamides (p. 25) • Quinolones (p. 26) • Polypeptides antibiotics (p. 27) • Glycopeptide Antibiotics (p. 28) • Sulfonamides (p. 29) • Lipoglycopeptides (p. 30) • First Generation Cephalosporins (p. 31) • Cephalosporin Third Generation (p. 32) • Fourth-Generation Cephalosporins (p. 33) • Fifth Generation Cephalosporin’s (p. 34) • Tetracycline antibiotics (p. 35) Computational Antibiotics Antibiotics Covered (in alphabetical order) Amikacin (p. 36) Cefempidone (p. 98) Ceftizoxime (p. 159) Amoxicillin (p. 38) Cefepime (p. 100) Ceftobiprole (p. 161) Ampicillin (p. 40) Cefetamet (p. 102) Ceftoxide (p. 163) Arsphenamine (p. 42) Cefetrizole (p. 104) Ceftriaxone (p. 165) Azithromycin (p.44) Cefivitril (p. 106) Cefuracetime (p. 167) Aziocillin (p. 46) Cefixime (p. 108) Cefuroxime (p. 169) Aztreonam (p.48) Cefmatilen ( p. 110) Cefuzonam (p. 171) Bacampicillin (p. 50) Cefmetazole (p. 112) Cefalexin (p. 173) Bacitracin (p. 52) Cefodizime (p. 114) Chloramphenicol (p.175) Balofloxacin (p. 54) Cefonicid (p. 116) Cilastatin (p. 177) Carbenicillin (p. 56) Cefoperazone (p. 118) Ciprofloxacin (p. 179) Cefacetrile (p. 58) Cefoselis (p. 120) Clarithromycin (p. 181) Cefaclor (p. -

Human Microbiota Network: Unveiling Potential Crosstalk Between the Different Microbiota Ecosystems and Their Role in Health and Disease
nutrients Review Human Microbiota Network: Unveiling Potential Crosstalk between the Different Microbiota Ecosystems and Their Role in Health and Disease Jose E. Martínez †, Augusto Vargas † , Tania Pérez-Sánchez , Ignacio J. Encío , Miriam Cabello-Olmo * and Miguel Barajas * Biochemistry Area, Department of Health Science, Public University of Navarre, 31008 Pamplona, Spain; [email protected] (J.E.M.); [email protected] (A.V.); [email protected] (T.P.-S.); [email protected] (I.J.E.) * Correspondence: [email protected] (M.C.-O.); [email protected] (M.B.) † These authors contributed equally to this work. Abstract: The human body is host to a large number of microorganisms which conform the human microbiota, that is known to play an important role in health and disease. Although most of the microorganisms that coexist with us are located in the gut, microbial cells present in other locations (like skin, respiratory tract, genitourinary tract, and the vaginal zone in women) also play a significant role regulating host health. The fact that there are different kinds of microbiota in different body areas does not mean they are independent. It is plausible that connection exist, and different studies have shown that the microbiota present in different zones of the human body has the capability of communicating through secondary metabolites. In this sense, dysbiosis in one body compartment Citation: Martínez, J.E.; Vargas, A.; may negatively affect distal areas and contribute to the development of diseases. Accordingly, it Pérez-Sánchez, T.; Encío, I.J.; could be hypothesized that the whole set of microbial cells that inhabit the human body form a Cabello-Olmo, M.; Barajas, M. -

EMA/CVMP/158366/2019 Committee for Medicinal Products for Veterinary Use
Ref. Ares(2019)6843167 - 05/11/2019 31 October 2019 EMA/CVMP/158366/2019 Committee for Medicinal Products for Veterinary Use Advice on implementing measures under Article 37(4) of Regulation (EU) 2019/6 on veterinary medicinal products – Criteria for the designation of antimicrobials to be reserved for treatment of certain infections in humans Official address Domenico Scarlattilaan 6 ● 1083 HS Amsterdam ● The Netherlands Address for visits and deliveries Refer to www.ema.europa.eu/how-to-find-us Send us a question Go to www.ema.europa.eu/contact Telephone +31 (0)88 781 6000 An agency of the European Union © European Medicines Agency, 2019. Reproduction is authorised provided the source is acknowledged. Introduction On 6 February 2019, the European Commission sent a request to the European Medicines Agency (EMA) for a report on the criteria for the designation of antimicrobials to be reserved for the treatment of certain infections in humans in order to preserve the efficacy of those antimicrobials. The Agency was requested to provide a report by 31 October 2019 containing recommendations to the Commission as to which criteria should be used to determine those antimicrobials to be reserved for treatment of certain infections in humans (this is also referred to as ‘criteria for designating antimicrobials for human use’, ‘restricting antimicrobials to human use’, or ‘reserved for human use only’). The Committee for Medicinal Products for Veterinary Use (CVMP) formed an expert group to prepare the scientific report. The group was composed of seven experts selected from the European network of experts, on the basis of recommendations from the national competent authorities, one expert nominated from European Food Safety Authority (EFSA), one expert nominated by European Centre for Disease Prevention and Control (ECDC), one expert with expertise on human infectious diseases, and two Agency staff members with expertise on development of antimicrobial resistance . -
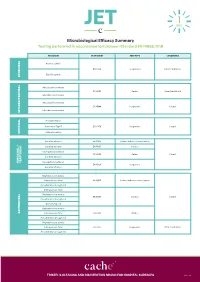
JET Microbiological Efficacy Summary
Microbiological Efficacy Summary Testing performed in accordance to European Standard EN 14885:2018 ORGANISM TEST NORM TEST TYPE CONDITIONS Bacillus subtilis EN 13704 Suspension Clean 1 and Dirty 1 Bacillus cereus SPORICIDAL Mycobacterium terrae EN 14563 Carrier Clean 1 and Dirty 2 Mycobacterium avium Mycobacterium terrae EN 14348 Suspension Clean 1 Mycobacterium avium MYCOBACTERICIDAL Poliovirus Type 1 Adenovirus Type 5 EN 14476 Suspension Clean 1 Murine Norovirus VIRUCIDAL Candida albicans EN 16615 Surface with mechanical action Candida albicans EN 13697 Surface Aspergillus brasiliensis EN 14562 Carrier Clean 1 Candida albicans YEASTICIDAL FUNGICIDAL / FUNGICIDAL Aspergillus brasiliensis EN 13624 Suspension Candida albicans Staphylococcus aureus Enterococcus hirae EN 16615 Surface with mechanical action Pseudomonas aeruginosa Enterococcus hirae Staphylococcus aureus EN 13697 Surface Clean 1 Pseudomonas aeruginosa Escherichia coli Staphylococcus aureus BACTERICIDAL Enterococcus hirae EN 14561 Carrier Pseudomonas aeruginosa Staphylococcus aureus Enterococcus hirae EN 13727 Suspension Clean 1 and Dirty 1 Pseudomonas aeruginosa TRISTEL’S CLEANING AND DISINFECTION BRAND FOR HOSPITAL SURFACES Page 1 of 3 Additional Testing TEST METHOD RNA DNA / Polyacrylamide gel electrophoresis (PAGE) ORGANISM TEST METHOD TEST TYPE CONDITIONS Acanthamoeba castellanii cysts Following the method of EN 13704 Suspension Clean 1 PROTOZOA Bacillus subtilis EN 17126 Suspension Clean 1 Bacillus cereus Clostridium difficile EN 13704 Suspension Clean 1 and Dirty 1 -

WHO Report on Surveillance of Antibiotic Consumption: 2016-2018 Early Implementation ISBN 978-92-4-151488-0 © World Health Organization 2018 Some Rights Reserved
WHO Report on Surveillance of Antibiotic Consumption 2016-2018 Early implementation WHO Report on Surveillance of Antibiotic Consumption 2016 - 2018 Early implementation WHO report on surveillance of antibiotic consumption: 2016-2018 early implementation ISBN 978-92-4-151488-0 © World Health Organization 2018 Some rights reserved. This work is available under the Creative Commons Attribution- NonCommercial-ShareAlike 3.0 IGO licence (CC BY-NC-SA 3.0 IGO; https://creativecommons. org/licenses/by-nc-sa/3.0/igo). Under the terms of this licence, you may copy, redistribute and adapt the work for non- commercial purposes, provided the work is appropriately cited, as indicated below. In any use of this work, there should be no suggestion that WHO endorses any specific organization, products or services. The use of the WHO logo is not permitted. If you adapt the work, then you must license your work under the same or equivalent Creative Commons licence. If you create a translation of this work, you should add the following disclaimer along with the suggested citation: “This translation was not created by the World Health Organization (WHO). WHO is not responsible for the content or accuracy of this translation. The original English edition shall be the binding and authentic edition”. Any mediation relating to disputes arising under the licence shall be conducted in accordance with the mediation rules of the World Intellectual Property Organization. Suggested citation. WHO report on surveillance of antibiotic consumption: 2016-2018 early implementation. Geneva: World Health Organization; 2018. Licence: CC BY-NC-SA 3.0 IGO. Cataloguing-in-Publication (CIP) data. -

A Genomic Journey Through a Genus of Large DNA Viruses
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Virology Papers Virology, Nebraska Center for 2013 Towards defining the chloroviruses: a genomic journey through a genus of large DNA viruses Adrien Jeanniard Aix-Marseille Université David D. Dunigan University of Nebraska-Lincoln, [email protected] James Gurnon University of Nebraska-Lincoln, [email protected] Irina V. Agarkova University of Nebraska-Lincoln, [email protected] Ming Kang University of Nebraska-Lincoln, [email protected] See next page for additional authors Follow this and additional works at: https://digitalcommons.unl.edu/virologypub Part of the Biological Phenomena, Cell Phenomena, and Immunity Commons, Cell and Developmental Biology Commons, Genetics and Genomics Commons, Infectious Disease Commons, Medical Immunology Commons, Medical Pathology Commons, and the Virology Commons Jeanniard, Adrien; Dunigan, David D.; Gurnon, James; Agarkova, Irina V.; Kang, Ming; Vitek, Jason; Duncan, Garry; McClung, O William; Larsen, Megan; Claverie, Jean-Michel; Van Etten, James L.; and Blanc, Guillaume, "Towards defining the chloroviruses: a genomic journey through a genus of large DNA viruses" (2013). Virology Papers. 245. https://digitalcommons.unl.edu/virologypub/245 This Article is brought to you for free and open access by the Virology, Nebraska Center for at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Virology Papers by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Authors Adrien Jeanniard, David D. Dunigan, James Gurnon, Irina V. Agarkova, Ming Kang, Jason Vitek, Garry Duncan, O William McClung, Megan Larsen, Jean-Michel Claverie, James L. Van Etten, and Guillaume Blanc This article is available at DigitalCommons@University of Nebraska - Lincoln: https://digitalcommons.unl.edu/ virologypub/245 Jeanniard, Dunigan, Gurnon, Agarkova, Kang, Vitek, Duncan, McClung, Larsen, Claverie, Van Etten & Blanc in BMC Genomics (2013) 14. -

(12) United States Patent (10) Patent No.: US 8,097,607 B2 Cabana Et Al
USO08097607B2 (12) United States Patent (10) Patent No.: US 8,097,607 B2 Cabana et al. (45) Date of Patent: *Jan. 17, 2012 (54) LOW DOSE RIFALAZIL COMPOSITIONS Emori et al., “Evaluation of in Vivo Therapeutic Efficacy of a New Benzoxazinorifamycin, KRM-1648, in SCID Mouse Model for Dis (76) Inventors: Bernard E. Cabana, Montgomery seminated Mycobacterium avium Complex Infection.” International Village, MD (US); Arthur F. Michaelis, Journal of Antimicrobial Agents 10(1):59 (1998). Devon, PA (US); Gary P. Magnant, Fujii et al., “In Vitro and In Vivo Antibacterial Activities of KRM Topsfield, MA (US); Chalom B. 1648 and KRM-1657, New Rifamycin Derivatives.” Antimicrobial Sayada, Luxembourg (LU) Agents and Chemotherapy 38: 1118, (1994). Gidoh et al., “Bactericidal Action at Low Doses of a New Rifamycin (*) Notice: Subject to any disclaimer, the term of this Derivative, 3'-hydroxy-5'-(4-isobutyl-1-piperazinyl) patent is extended or adjusted under 35 Benzoxazinorifamycin (KRM-1648) on Mycobacterium leprae U.S.C. 154(b) by 447 days. Inoculated into Ffootpads of Nude Mice.” Leprosy Review 63(4):319 This patent is Subject to a terminal dis (1992). claimer. Heep et al., “Detection of Rifabutin Resistance and Association of rpoB Mutation S with Resistance to Four Rifamycin Derivatives in Helicobacter pylori.” Journal of Clinical Microbiology & Infectious (21) Appl. No.: 10/668,792 Diseases 21:143 (2002). Hirara et al., “In Vitro and in Vivo Activities of the (22) Filed: Sep. 23, 2003 Benezoxazinorifamycin KRM-1648 Against Mycobacterium tuber (65) Prior Publication Data culosis,” Antimocrobial Agents and Chemotherapy 39 (10):2295 (1995). US 2004/O15784.0 A1 Aug. -

Pharmaceuticals As Environmental Contaminants
PharmaceuticalsPharmaceuticals asas EnvironmentalEnvironmental Contaminants:Contaminants: anan OverviewOverview ofof thethe ScienceScience Christian G. Daughton, Ph.D. Chief, Environmental Chemistry Branch Environmental Sciences Division National Exposure Research Laboratory Office of Research and Development Environmental Protection Agency Las Vegas, Nevada 89119 [email protected] Office of Research and Development National Exposure Research Laboratory, Environmental Sciences Division, Las Vegas, Nevada Why and how do drugs contaminate the environment? What might it all mean? How do we prevent it? Office of Research and Development National Exposure Research Laboratory, Environmental Sciences Division, Las Vegas, Nevada This talk presents only a cursory overview of some of the many science issues surrounding the topic of pharmaceuticals as environmental contaminants Office of Research and Development National Exposure Research Laboratory, Environmental Sciences Division, Las Vegas, Nevada A Clarification We sometimes loosely (but incorrectly) refer to drugs, medicines, medications, or pharmaceuticals as being the substances that contaminant the environment. The actual environmental contaminants, however, are the active pharmaceutical ingredients – APIs. These terms are all often used interchangeably Office of Research and Development National Exposure Research Laboratory, Environmental Sciences Division, Las Vegas, Nevada Office of Research and Development Available: http://www.epa.gov/nerlesd1/chemistry/pharma/image/drawing.pdfNational -

Sample Report Vaginosis.Pdf
LAB #: Sample Report CLIENT #: 12345 PATIENT: Sample Patient DOCTOR: Sample Doctor ID: Doctor's Data, Inc. SEX: Female 3755 Illinois Ave. DOB: 01/01/1993 AGE: 25 St. Charles, IL 60174 U.S.A. !"#$%&'(%(!)*'+%,- GRAM STAIN MICROSCOPY BACTERIAL VAGINOSIS SCORE Normal Abnormal Expected score interpretation: Lactobacilli None Mod - Many 0 - 3 BV not likely 4 - 6 BV indeterminate Curved Gram 10 7-10 BV highly suggestive Negative Rods Many None Small Gram 1 Many None The BV score is calculated based upon the gram stain results Negative Rods and is independent of the yeast, and bacterial cultures. 1Nugent Scoring System. (Nugent et al. J. Clin. Micro. Yeast None None (1991)29:297-301) RBC’s None None YEAST CULTURE 2+ Candida albicans WBC’s 0 0 - 6 Clue Cells Present None Eosinophils N/A None Eosinophils reported and Wrights Stain performed when WBC’s >6 Additional Gram Stain Findings: Rare Gram positive cocci in pairs BACTERIOLOGY CULTURE Expected/Beneficial flora Commensal (Imbalanced) flora Dysbiotic flora NG Lactobacillus spp. 2+ Alpha hemolytic strep 3+ Gardnerella vaginalis 2+ Gamma hemolytic strep 4+ Enterococcus faecalis 1+ Staphylococcus not aureus NG = No Growth SPECIMEN DATA Comments: Date Collected: 01/14/2019 Date Received: 01/16/2019 Date Completed: 01/23/2019 ©Doctor’s Data, Inc. !!! ADDRESS: 3755 Illinois Avenue, St. Charles, IL 60174-2420 !!! MED DIR: Erlo Roth, MD !!! CLIA ID NO: 14D0646470 0002038 LAB #: Sample Report CLIENT #: 12345 PATIENT: Sample DOCTOR: Sample Doctor Patient ID: Doctor's Data, Inc. SEX: Female 3755 Illinois Ave. DOB: 01/01/1993 St. Charles, IL 60174 U.S.A. -

Mycobacterium Avium Possesses Extracellular DNA That Contributes to Biofilm Formation, Structural Integrity, and Tolerance to Antibiotics
Mycobacterium avium Possesses Extracellular DNA that Contributes to Biofilm Formation, Structural Integrity, and Tolerance to Antibiotics Rose, S. J., Babrak, L. M., & Bermudez, L. E. (2015). Mycobacterium avium Possesses Extracellular DNA that Contributes to Biofilm Formation, Structural Integrity, and Tolerance to Antibiotics. PLoS ONE, 10(5), e0128772. doi: 10.1371/journal.pone.0128772 10.1371/journal.pone.0128772 Public Library of Science Version of Record http://cdss.library.oregonstate.edu/sa-termsofuse RESEARCH ARTICLE Mycobacterium avium Possesses Extracellular DNA that Contributes to Biofilm Formation, Structural Integrity, and Tolerance to Antibiotics Sasha J. Rose1,2, Lmar M. Babrak1,2, Luiz E. Bermudez1,2* 1 Department of Biomedical Sciences, College of Veterinary Medicine, Oregon State University, Corvallis, Oregon, United States of America, 2 Department of Microbiology, College of Science, Oregon State University, Corvallis, Oregon, United States of America * [email protected] Abstract Mycobacterium avium subsp. hominissuis is an opportunistic pathogen that is associated with biofilm-related infections of the respiratory tract and is difficult to treat. In recent years, extracellular DNA (eDNA) has been found to be a major component of bacterial biofilms, in- OPEN ACCESS cluding many pathogens involved in biofilm-associated infections. To date, eDNA has not Citation: Rose SJ, Babrak LM, Bermudez LE (2015) been described as a component of mycobacterial biofilms. In this study, we identified and Mycobacterium avium Possesses Extracellular DNA characterized eDNA in a high biofilm-producing strain of Mycobacterium avium subsp. that Contributes to Biofilm Formation, Structural hominissuis (MAH). In addition, we surveyed for presence of eDNA in various MAH strains Integrity, and Tolerance to Antibiotics. -

Surveillance of Antimicrobial Consumption in Europe 2013-2014 SURVEILLANCE REPORT
SURVEILLANCE REPORT SURVEILLANCE REPORT Surveillance of antimicrobial consumption in Europe in Europe consumption of antimicrobial Surveillance Surveillance of antimicrobial consumption in Europe 2013-2014 2012 www.ecdc.europa.eu ECDC SURVEILLANCE REPORT Surveillance of antimicrobial consumption in Europe 2013–2014 This report of the European Centre for Disease Prevention and Control (ECDC) was coordinated by Klaus Weist. Contributing authors Klaus Weist, Arno Muller, Ana Hoxha, Vera Vlahović-Palčevski, Christelle Elias, Dominique Monnet and Ole Heuer. Data analysis: Klaus Weist, Arno Muller and Ana Hoxha. Acknowledgements The authors would like to thank the ESAC-Net Disease Network Coordination Committee members (Marcel Bruch, Philippe Cavalié, Herman Goossens, Jenny Hellman, Susan Hopkins, Stephanie Natsch, Anna Olczak-Pienkowska, Ajay Oza, Arjana Tambić Andrasevic, Peter Zarb) and observers (Jane Robertson, Arno Muller, Mike Sharland, Theo Verheij) for providing valuable comments and scientific advice during the production of the report. All ESAC-Net participants and National Coordinators are acknowledged for providing data and valuable comments on this report. The authors also acknowledge Gaetan Guyodo, Catalin Albu and Anna Renau-Rosell for managing the data and providing technical support to the participating countries. Suggested citation: European Centre for Disease Prevention and Control. Surveillance of antimicrobial consumption in Europe, 2013‒2014. Stockholm: ECDC; 2018. Stockholm, May 2018 ISBN 978-92-9498-187-5 ISSN 2315-0955