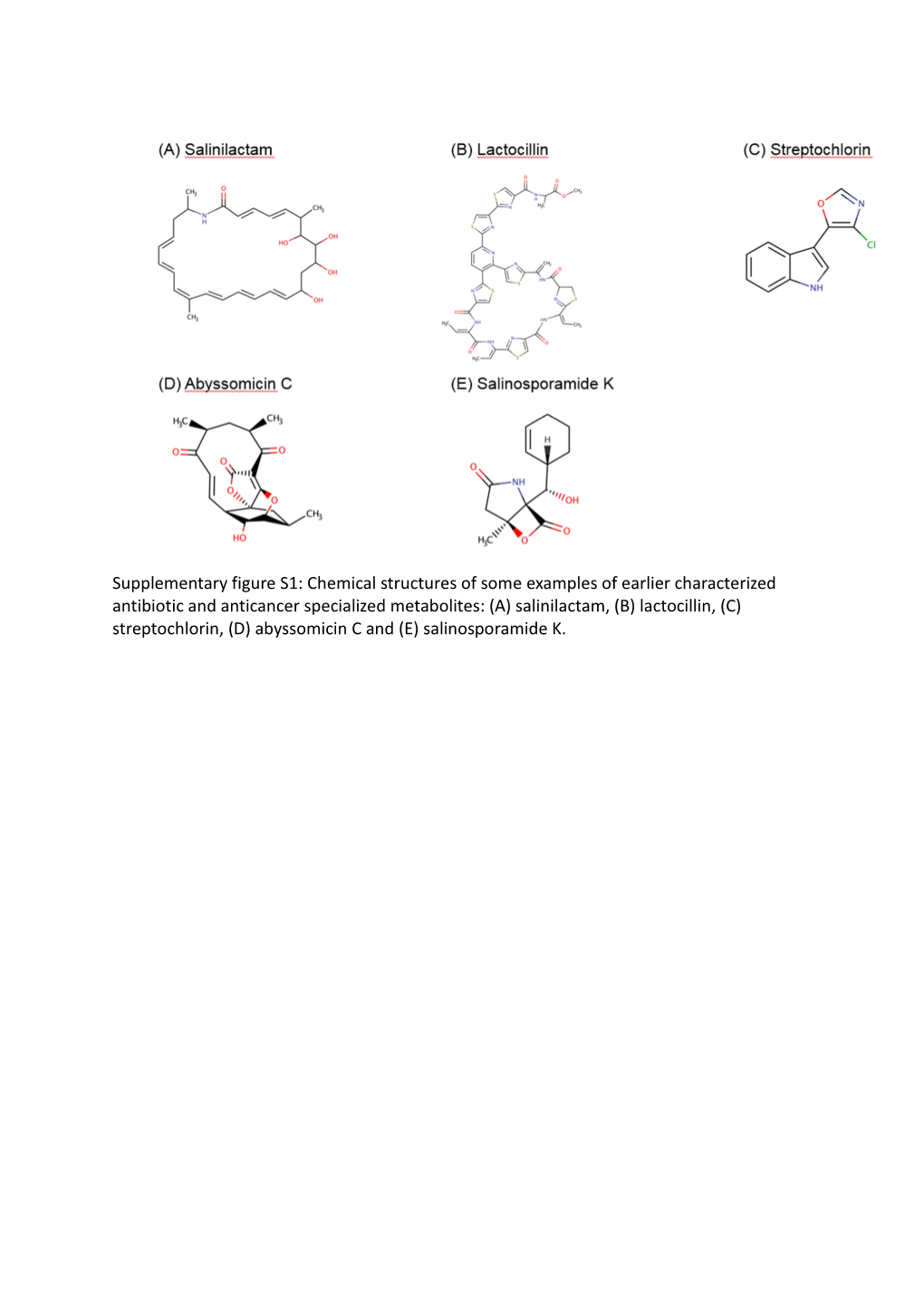
Chemical Structures of Some Examples of Earlier Characterized Antibiotic and Anticancer Specialized
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Asaia Bogorensis Gen. Nov., Sp. Nov., an Unusual Acetic Acid Bacterium in the Α-Proteobacteria
International Journal of Systematic and Evolutionary Microbiology (2000) 50, 823–829 Printed in Great Britain Asaia bogorensis gen. nov., sp. nov., an unusual acetic acid bacterium in the α-Proteobacteria Yuzo Yamada,1 Kazushige Katsura,1 Hiroko Kawasaki,2 Yantyati Widyastuti,3 Susono Saono,3 Tatsuji Seki,2 Tai Uchimura1 and Kazuo Komagata1 Author for correspondence: Yuzo Yamada. Tel\Fax: j81 54 635 2316. 1 Laboratory of General and Eight Gram-negative, aerobic, rod-shaped and peritrichously flagellated strains Applied Microbiology, were isolated from flowers of the orchid tree (Bauhinia purpurea) and of Department of Applied Biology and Chemistry, plumbago (Plumbago auriculata), and from fermented glutinous rice, all Faculty of Applied collected in Indonesia. The enrichment culture approach for acetic acid bacteria Bioscience, Tokyo was employed, involving use of sorbitol medium at pH 35. All isolates grew University of Agriculture, Sakuragaoka 1-1-1, well at pH 30 and 30 SC. They did not oxidize ethanol to acetic acid except for Setagaya-ku, one strain that oxidized ethanol weakly, and 035% acetic acid inhibited their Tokoyo 156-8502, Japan growth completely. However, they oxidized acetate and lactate to carbon 2 The International Center dioxide and water. The isolates grew well on mannitol agar and on glutamate for Biotechnology, Osaka agar, and assimilated ammonium sulfate for growth on vitamin-free glucose University, Yamadaoka 2-1, Suita, Osaka 565-0871, medium. The isolates produced acid from D-glucose, D-fructose, L-sorbose, Japan dulcitol and glycerol. The quinone system was Q-10. DNA base composition 3 Research and ranged from 593to610 mol% GMC. -

The 2014 Golden Gate National Parks Bioblitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event
National Park Service U.S. Department of the Interior Natural Resource Stewardship and Science The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 ON THIS PAGE Photograph of BioBlitz participants conducting data entry into iNaturalist. Photograph courtesy of the National Park Service. ON THE COVER Photograph of BioBlitz participants collecting aquatic species data in the Presidio of San Francisco. Photograph courtesy of National Park Service. The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 Elizabeth Edson1, Michelle O’Herron1, Alison Forrestel2, Daniel George3 1Golden Gate Parks Conservancy Building 201 Fort Mason San Francisco, CA 94129 2National Park Service. Golden Gate National Recreation Area Fort Cronkhite, Bldg. 1061 Sausalito, CA 94965 3National Park Service. San Francisco Bay Area Network Inventory & Monitoring Program Manager Fort Cronkhite, Bldg. 1063 Sausalito, CA 94965 March 2016 U.S. Department of the Interior National Park Service Natural Resource Stewardship and Science Fort Collins, Colorado The National Park Service, Natural Resource Stewardship and Science office in Fort Collins, Colorado, publishes a range of reports that address natural resource topics. These reports are of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Report Series is used to disseminate comprehensive information and analysis about natural resources and related topics concerning lands managed by the National Park Service. -

Indoor Microbiome, Environmental Characteristics and Asthma Among Junior High
Indoor microbiome, environmental characteristics and asthma among junior high school students in Johor Bahru, Malaysia Xi Fu1, Dan Norbäck2, Qianqian Yuan3,4, Yanling Li3,4, Xunhua Zhu3,4,Yiqun Deng3,4, Jamal Hisham Hashim5,6, Zailina Hashim7, Yi-Wu Zheng8, Xu-Xin Lai8, Michael Dho Spangfort8, Yu Sun3,4 * 1Department of Occupational and Environmental Health, School of Public Health, Sun Yat-sen University, Guangzhou, PR China. 2Occupational and Environmental Medicine, Dept. of Medical Science, University Hospital, Uppsala University, 75237 Uppsala, Sweden. 3Guangdong Provincial Key Laboratory of Protein Function and Regulation in Agricultural Organisms, College of Life Sciences, South China Agricultural University, Guangzhou, Guangdong, 510642, PR China. 4Key Laboratory of Zoonosis of Ministry of Agriculture and Rural Affairs, South China Agricultural University, Guangzhou, Guangdong, 510642, PR China. 5United Nations University-International Institute for Global Health, Kuala Lumpur, Malaysia, 6Department of Community Health, National University of Malaysia, Kuala Lumpur, Malaysia 7Department of Environmental and Occupational Health, Faculty of Medicine and Health Sciences, Universiti Putra Malaysia, UPM, Serdang, Selangor, Malaysia 8Asia Pacific Research, ALK-Abello A/S, Guanzhou, China *To whom corresponds should be addressed: Yu Sun [email protected], Key words: bacteria, fungi, microbial community, absolute quantity, wheezing, breathlessness, adolescence, dampness/visible mold Abstract Indoor microbial diversity and composition are suggested to affect the prevalence and severity of asthma, but no microbial association study has been conducted in tropical countries. In this study, we collected floor dust and environmental characteristics from 21 classrooms, and health data related to asthma symptoms from 309 students, in junior high schools in Johor Bahru, Malaysia. -

Methanogenic Activity in Río Tinto, a Terrestrial Mars Analogue R
Methanogenic activity in Río Tinto, a terrestrial Mars analogue R. Amils Centro de Biología Molecular Severo Ochoa (UAM-CSIC) y Centro de Astrobiología (INTA- CSIC) Frascati, noviembre 2009 new insides in the Mars exploration H2O on Mars methane (PFS) it can be concluded that on Mars there are sedimentary rocks that were formed in acidic conditions (acidic lakes or oceans) • possible terrestrial analogs: - submarine hydrothermalism - acidic environments to explore the deep sea requires expensive equipment (Alvin) natural acidic waters natural acidic environments: - areas with volcanic activity 0 SO2 + H2S ——> S + H2O - metal mining activities 3+ 2- + FeS2 + H2O —> Fe + SO4 + H in this case the extreme acidic conditions are promoted by biological activity geomicrobiology of metallic sulfides pyrite, molibdenite, tungstenite (thiosulfate mec.) 3+ 2- 2+ + FeS2+6Fe +3H2O → S2O3 +7Fe +6H 2- 3+ 2- 2+ + S2O3 +8Fe +5H2O → 2SO4 +8Fe +10H Rest of sulfides (polisulfide mec.) 3+ + 2+ 2+ 8MS+8Fe +8H → 8M +4H2Sn+8Fe (n≥2) 3+ o 2+ + 4H2Sn+8Fe → S8 +8Fe +8H o 2- + S8 +4H2O (S oxidizers) → SO4 +8H Bacterias come-meteoritos role of the microbial activity in the leaching of pyrite chemical 3+ reaction Fe Fe2+ microbial activity 2- + SO4 + H Rio Tinto rise at the heart of the Iberian Pyritic Belt Río Tinto is an acidic river, pH 2.3, 100 km long and with a high concentration of soluble metals the iron concentration at the origin is between 15-20 g/l and the sulfate is constant and around 15 g/l geoMICROBIOLOGY combination of conventional microbial ecology techniques and molecular ecology tools A B Phylogeny of acidophilic microorganisms detected in Rio Tinto Actinobacteria Cyanobacteria . -

MICROBIOLOGY ECOLOGY MICROBIOLOGY (Bjarnason, 1988)
RESEARCH ARTICLE Analysis of the unique geothermal microbial ecosystem of the Blue Lagoon Solveig K. Petursdottir,1 Snaedis H. Bjornsdottir,1,2 Gudmundur O. Hreggvidsson,1,2 Sigridur Hjorleifsdottir1 & Jakob K. Kristjansson3 1Prokaria-Matis Ltd, Reykjavik, Iceland; 2Institute of Biology, University of Iceland, Reykjavik, Iceland; and 3Arkea Ltd, Reykajvik, Iceland Correspondence: Solveig K. Petursdottir, Abstract Prokaria-Matis Ltd, Gylfaflot¨ 5, IS-112, Reykjavik, Iceland. Tel.: 1354 858 5123; fax: Cultivation and culture-independent techniques were used to describe the geother- 1354 422 5002; e-mail: [email protected] mal ecosystem of the Blue Lagoon in Iceland. The lagoon contains both seawater Downloaded from and freshwater of geothermal origin and is extremely high in silica content. Water Received 16 October 2008; revised 11 April samples were collected repeatedly in summer and autumn in 2003 and 2005 and in 2009; accepted 27 July 2009. winter 2006 were analyzed for species composition. The study revealed the typical Final version published online 26 August 2009. traits of an extreme ecosystem characterized by dominating species and other species represented in low numbers. A total of 35 taxa were identified. The DOI:10.1111/j.1574-6941.2009.00757.x calculated biodiversity index of the samples was 2.1–2.5. The majority (83%) of http://femsec.oxfordjournals.org/ analyzed taxa were closely related to bacteria of marine and geothermal origin Editor: Riks Laanbroek reflecting a marine character of the ecosystem and the origin of the Blue Lagoon hydrothermal fluid. A high ratio (63%) of analyzed taxa represented putative novel Keywords ecosystem; geothermal; biodiversity; bacterial species. -

Microbial Community Structure Dynamics in Ohio River Sediments During Reductive Dechlorination of Pcbs
University of Kentucky UKnowledge University of Kentucky Doctoral Dissertations Graduate School 2008 MICROBIAL COMMUNITY STRUCTURE DYNAMICS IN OHIO RIVER SEDIMENTS DURING REDUCTIVE DECHLORINATION OF PCBS Andres Enrique Nunez University of Kentucky Right click to open a feedback form in a new tab to let us know how this document benefits ou.y Recommended Citation Nunez, Andres Enrique, "MICROBIAL COMMUNITY STRUCTURE DYNAMICS IN OHIO RIVER SEDIMENTS DURING REDUCTIVE DECHLORINATION OF PCBS" (2008). University of Kentucky Doctoral Dissertations. 679. https://uknowledge.uky.edu/gradschool_diss/679 This Dissertation is brought to you for free and open access by the Graduate School at UKnowledge. It has been accepted for inclusion in University of Kentucky Doctoral Dissertations by an authorized administrator of UKnowledge. For more information, please contact [email protected]. ABSTRACT OF DISSERTATION Andres Enrique Nunez The Graduate School University of Kentucky 2008 MICROBIAL COMMUNITY STRUCTURE DYNAMICS IN OHIO RIVER SEDIMENTS DURING REDUCTIVE DECHLORINATION OF PCBS ABSTRACT OF DISSERTATION A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the College of Agriculture at the University of Kentucky By Andres Enrique Nunez Director: Dr. Elisa M. D’Angelo Lexington, KY 2008 Copyright © Andres Enrique Nunez 2008 ABSTRACT OF DISSERTATION MICROBIAL COMMUNITY STRUCTURE DYNAMICS IN OHIO RIVER SEDIMENTS DURING REDUCTIVE DECHLORINATION OF PCBS The entire stretch of the Ohio River is under fish consumption advisories due to contamination with polychlorinated biphenyls (PCBs). In this study, natural attenuation and biostimulation of PCBs and microbial communities responsible for PCB transformations were investigated in Ohio River sediments. Natural attenuation of PCBs was negligible in sediments, which was likely attributed to low temperature conditions during most of the year, as well as low amounts of available nitrogen, phosphorus, and organic carbon. -

Identification of Genes Responsible for Ochratoxin-A Biodegradation By
Szent István University PhD School of Environmental Sciences Identification of genes responsible for ochratoxin-A biodegradation by Cupriavidus basilensis ŐR16 and valuation of Cupriavidus genus for mycotoxins biodegradation potential Thesis of Doctoral Dissertation (PhD) Mohammed Talib Jasim AL-Nussairawi Gödöllő, Hungary 2020 I. DECLARATION I declare that this thesis is a record of original work and contains no material that has been accepted for the award of any other degree or diploma in any university. To the best of my knowledge and belief, this thesis contains no material previously published or written by another person, except where due reference is made in the text. Name: Ph.D. School of Environmental Sciences Discipline: Environmental Sciences / Biotechnology Leader of Doctoral School: Csákiné Dr. Erika Michéli, Ph.D. professor, head of department Department of Soil science and Agrochemistry Faculty of Agricultural and Environmental Sciences Institute of Environmental Sciences Szent István University Supervisor: Matyas Cserhati, Ph.D. associate professor Department of Environmental Protection and Ecotoxicology Faculty of Agricultural and Environmental Sciences Institute of Aquaculture and Environmental Protection Szent István University ........................................................... ................................................... Approval of School Leader Approval of Supervisor 2 Table of Contents I. DECLARATION ................................................................................................................... -

Annual Conference Abstracts
ANNUAL CONFERENCE 14-17 April 2014 Arena and Convention Centre, Liverpool ABSTRACTS SGM ANNUAL CONFERENCE APRIL 2014 ABSTRACTS (LI00Mo1210) – SGM Prize Medal Lecture (LI00Tu1210) – Marjory Stephenson Climate Change, Oceans, and Infectious Disease Prize Lecture Dr. Rita R. Colwell Understanding the basis of antibiotic resistance University of Maryland, College Park, MD, USA as a platform for early drug discovery During the mid-1980s, satellite sensors were developed to monitor Laura JV Piddock land and oceans for purposes of understanding climate, weather, School of Immunity & Infection and Institute of Microbiology and and vegetation distribution and seasonal variations. Subsequently Infection, University of Birmingham, UK inter-relationships of the environment and infectious diseases Antibiotic resistant bacteria are one of the greatest threats to human were investigated, both qualitatively and quantitatively, with health. Resistance can be mediated by numerous mechanisms documentation of the seasonality of diseases, notably malaria including mutations conferring changes to the genes encoding the and cholera by epidemiologists. The new research revealed a very target proteins as well as RND efflux pumps, which confer innate close interaction of the environment and many other infectious multi-drug resistance (MDR) to bacteria. The production of efflux diseases. With satellite sensors, these relationships were pumps can be increased, usually due to mutations in regulatory quantified and comparatively analyzed. More recent studies of genes, and this confers resistance to antibiotics that are often used epidemic diseases have provided models, both retrospective and to treat infections by Gram negative bacteria. RND MDR efflux prospective, for understanding and predicting disease epidemics, systems not only confer antibiotic resistance, but altered expression notably vector borne diseases. -

“Candidatus Deianiraea Vastatrix” with the Ciliate Paramecium Suggests
bioRxiv preprint doi: https://doi.org/10.1101/479196; this version posted November 27, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. The extracellular association of the bacterium “Candidatus Deianiraea vastatrix” with the ciliate Paramecium suggests an alternative scenario for the evolution of Rickettsiales 5 Castelli M.1, Sabaneyeva E.2, Lanzoni O.3, Lebedeva N.4, Floriano A.M.5, Gaiarsa S.5,6, Benken K.7, Modeo L. 3, Bandi C.1, Potekhin A.8, Sassera D.5*, Petroni G.3* 1. Centro Romeo ed Enrica Invernizzi Ricerca Pediatrica, Dipartimento di Bioscienze, Università 10 degli studi di Milano, Milan, Italy 2. Department of Cytology and Histology, Faculty of Biology, Saint Petersburg State University, Saint-Petersburg, Russia 3. Dipartimento di Biologia, Università di Pisa, Pisa, Italy 4 Centre of Core Facilities “Culture Collections of Microorganisms”, Saint Petersburg State 15 University, Saint Petersburg, Russia 5. Dipartimento di Biologia e Biotecnologie, Università degli studi di Pavia, Pavia, Italy 6. UOC Microbiologia e Virologia, Fondazione IRCCS Policlinico San Matteo, Pavia, Italy 7. Core Facility Center for Microscopy and Microanalysis, Saint Petersburg State University, Saint- Petersburg, Russia 20 8. Department of Microbiology, Faculty of Biology, Saint Petersburg State University, Saint- Petersburg, Russia * Corresponding authors, contacts: [email protected] ; [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/479196; this version posted November 27, 2018. -

Motiliproteus Sediminis Gen. Nov., Sp. Nov., Isolated from Coastal Sediment
Antonie van Leeuwenhoek (2014) 106:615–621 DOI 10.1007/s10482-014-0232-2 ORIGINAL PAPER Motiliproteus sediminis gen. nov., sp. nov., isolated from coastal sediment Zong-Jie Wang • Zhi-Hong Xie • Chao Wang • Zong-Jun Du • Guan-Jun Chen Received: 3 April 2014 / Accepted: 4 July 2014 / Published online: 20 July 2014 Ó Springer International Publishing Switzerland 2014 Abstract A novel Gram-stain-negative, rod-to- demonstrated that the novel isolate was 93.3 % similar spiral-shaped, oxidase- and catalase- positive and to the type strain of Neptunomonas antarctica, 93.2 % facultatively aerobic bacterium, designated HS6T, was to Neptunomonas japonicum and 93.1 % to Marino- isolated from marine sediment of Yellow Sea, China. bacterium rhizophilum, the closest cultivated rela- It can reduce nitrate to nitrite and grow well in marine tives. The polar lipid profile of the novel strain broth 2216 (MB, Hope Biol-Technology Co., Ltd) consisted of phosphatidylethanolamine, phosphatidyl- with an optimal temperature for growth of 30–33 °C glycerol and some other unknown lipids. Major (range 12–45 °C) and in the presence of 2–3 % (w/v) cellular fatty acids were summed feature 3 (C16:1 NaCl (range 0.5–7 %, w/v). The pH range for growth x7c/iso-C15:0 2-OH), C18:1 x7c and C16:0 and the main was pH 6.2–9.0, with an optimum at 6.5–7.0. Phylo- respiratory quinone was Q-8. The DNA G?C content genetic analysis based on 16S rRNA gene sequences of strain HS6T was 61.2 mol %. Based on the phylogenetic, physiological and biochemical charac- teristics, strain HS6T represents a novel genus and The GenBank accession number for the 16S rRNA gene T species and the name Motiliproteus sediminis gen. -
The RDP-II Backbone Tree for Release 8.0. the Tree Was Inferred from a Distance Matrix Generated in PAUP* with the Weighbor (Weighted Neighbor Joining) Algorithm
Methylococcus capsulatus ACM 1292 (T) Oceanospirillum linum ATCC 11336 (T) Halomonas halodenitrificans ATCC 13511 (T) Legionella lytica PCM 2298 (T) Francisella tularensis subsp. tularensis ATCC 6223 (T) Coxiella burnetii Q177 Moraxella catarrhalis ATCC 25238 (T) Pseudomonas fluorescens IAM 12022 (T) Piscirickettsia salmonis LF-89 (T) Thiothrix nivea DSM 5205 (T) Allochromatium minutissimum DSM 1376 (T) Alteromonas macleodii IAM 12920 (T) Aeromonas salmonicida subsp. smithia CCM 4103 (T) Pasteurella multocida NCTC 10322 (T) Enterobacter nimipressuralis LMG 10245 (T) Vibrio vulnificus ATCC 27562 (T) Ectothiorhodospira mobilis DSM237 (T) Xanthomonas campestris LMG 568 (T) Cardiobacterium hominis ATCC 15826 (T) Methylophilus methylotrophus ATCC 53528 (T) Rhodocyclus tenuis DSM 109 (T) Hydrogenophilus thermoluteolus TH-1 (T) Neisseria gonorrhoeae NCTC 8375 (T) Comamonas testosteroni ATCC 11996 (T) Nitrosospira multiformis ATCC 25196 (T) Spirillum volutans ATCC 19554 (T) Burkholderia glathei LMG 14190 (T) Alcaligenes defragrans DSM 12141 (T) Oxalobacter formigenes ATCC 35274 (T) Acetobacter oboediens DSM 11826 (T) clone CS93 PROTEOBACTERIA Caedibacter caryophilus 221 (T) Rhodobacter sphaeroides ATCC 17023 (T) Rickettsia rickettsii ATCC VR-891 (T) Ehrlichia risticii ATCC VR-986 (T) Sphingomonas paucimobilis GIFU 2395 (T) Caulobacter fusiformis ATCC 15257 (T) Rhodospirillum rubrum ATCC 11170 (T) Brucella melitensis ATCC 23459 (T) Rhizobium tropici IFO 15247 (T) Bartonella vinsonii subsp. vinsonii ATCC VR-152 (T) Phyllobacterium myrsinacearum -

Geomicrobiological Processes in Extreme Environments: a Review
202 Articles by Hailiang Dong1, 2 and Bingsong Yu1,3 Geomicrobiological processes in extreme environments: A review 1 Geomicrobiology Laboratory, China University of Geosciences, Beijing, 100083, China. 2 Department of Geology, Miami University, Oxford, OH, 45056, USA. Email: [email protected] 3 School of Earth Sciences, China University of Geosciences, Beijing, 100083, China. The last decade has seen an extraordinary growth of and Mancinelli, 2001). These unique conditions have selected Geomicrobiology. Microorganisms have been studied in unique microorganisms and novel metabolic functions. Readers are directed to recent review papers (Kieft and Phelps, 1997; Pedersen, numerous extreme environments on Earth, ranging from 1997; Krumholz, 2000; Pedersen, 2000; Rothschild and crystalline rocks from the deep subsurface, ancient Mancinelli, 2001; Amend and Teske, 2005; Fredrickson and Balk- sedimentary rocks and hypersaline lakes, to dry deserts will, 2006). A recent study suggests the importance of pressure in the origination of life and biomolecules (Sharma et al., 2002). In and deep-ocean hydrothermal vent systems. In light of this short review and in light of some most recent developments, this recent progress, we review several currently active we focus on two specific aspects: novel metabolic functions and research frontiers: deep continental subsurface micro- energy sources. biology, microbial ecology in saline lakes, microbial Some metabolic functions of continental subsurface formation of dolomite, geomicrobiology in dry deserts, microorganisms fossil DNA and its use in recovery of paleoenviron- Because of the unique geochemical, hydrological, and geological mental conditions, and geomicrobiology of oceans. conditions of the deep subsurface, microorganisms from these envi- Throughout this article we emphasize geomicrobiological ronments are different from surface organisms in their metabolic processes in these extreme environments.