A Cross-Sectional Epidemiological Study of Domestic Animals Related to Human
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Demobilizing and Integrating the Nicaraguan Resistance 1990-1997
The International Commission for Support and Verification Commission (CIAV) Demobilizing and Integrating the Nicaraguan Resistance 1990-1997 ii Acknowledgements: This paper is a summary English version, written by Fernando Arocena, a consultant to CIAV-OAS, based on the original Spanish report: “La Comisión Internacional de Apoyo y Verificación, La Desmovilización y Reinserción de la Resistencia Nicaragüense 1990 – 1997”, prepared by Héctor Vanolli, Diógenes Ruiz and Arturo Wallace, also consultants to the CIAV-OAS. Bruce Rickerson, Senior Specialist at the UPD revised and edited the English text. This is a publication of the General Secretariat of the Organization of American States. The ideas, thoughts, and opinions expressed are not necessarily those of the OAS or its member states. The opinions expressed are the responsibility of the authors. Correspondence should be directed to the UPD, 1889 "F" Street, N.W., 8th Floor, Washington, DC, 20006, USA. Copyright ©1998 by OAS. All rights reserved. This publication may be reproduced provided credit is given to the source. TABLE OF CONTENTS ACRONYMS................................................................................................................................ix READER'S GUIDE ..................................................................................................................... xi INTRODUCTION......................................................................................................................xiii EXECUTIVE SUMMARY ....................................................................................................... -

Infected Areas As at 11 May 1995 Zones Infectées Au 11 Mai 1995
WEEKLY EPIDEMIOLOGICAL RECORD, Ho. It, 12 MAY 1995 • RELEVÉ ÉPIDÉMIOLOGIQUE HEBDOMADAIRE, N‘ H , 12 MAI 1995 M adagascar (4 May 1995).1 The number of influenza M adagascar (4 mai 1995).1 Le nombre d’isolements de virus A(H3N2) virus isolates increased during February and grippaux A(H3N2) s’est accru en février et en mars. Un accrois March. At that time there was a noticeable increase in sement marqué des syndromes grippaux a alors été observé parmi influenza-like illness among the general population in la population générale à Antananarivo. Des virus grippaux Antananarivo. Influenza A(H3N2) viruses continued to be A(H3N2) ont continué à être isolés en avril, de même que quel isolated in April along with a few of H1N1 subtype. ques virus appartenant au sous-type H1N1. Norway (3 May 1995).2 The notifications of influenza-like Norvège (3 mai 1995).2 Les notifications de syndrome grippal ont illness reached a peak in the last week of March and had atteint un pic la dernière semaine de mars et sont retombées à 89 declined to 89 per 100 000 population in the week ending pour 100 000 habitants au cours de la semaine qui s’est achevée le 23 April. At that time, 7 counties, mainly in the south-east 23 avril. Sept comtés, principalement dans le sud-est et l’ouest du and the west, reported incidence rates above 100 per pays, signalaient alors des taux d ’incidence dépassant 100 pour 100 000 and in the following week, 4 counties reported 100 000, et la semaine suivante 4 comtés ont déclaré des taux au- rates above 100. -

WEEKLY EPIDEMIOLOGICAL RECORD RELEVE EPIDEMIOLOGIQUE HEBDOMADAIRE 15 SEPTEMBER 1995 ● 70Th YEAR 70E ANNÉE ● 15 SEPTEMBRE 1995
WEEKLY EPIDEMIOLOGICAL RECORD, No. 37, 15 SEPTEMBER 1995 • RELEVÉ ÉPIDÉMIOLOGIQUE HEBDOMADAIRE, No 37, 15 SEPTEMBRE 1995 1995, 70, 261-268 No. 37 World Health Organization, Geneva Organisation mondiale de la Santé, Genève WEEKLY EPIDEMIOLOGICAL RECORD RELEVE EPIDEMIOLOGIQUE HEBDOMADAIRE 15 SEPTEMBER 1995 c 70th YEAR 70e ANNÉE c 15 SEPTEMBRE 1995 CONTENTS SOMMAIRE Expanded Programme on Immunization – Programme élargi de vaccination – Lot Quality Assurance Evaluation de la couverture vaccinale par la méthode dite de Lot survey to assess immunization coverage, Quality Assurance (échantillonnage par lots pour l'assurance de la qualité), Burkina Faso 261 Burkina Faso 261 Human rabies in the Americas 264 La rage humaine dans les Amériques 264 Influenza 266 Grippe 266 List of infected areas 266 Liste des zones infectées 266 Diseases subject to the Regulations 268 Maladies soumises au Règlement 268 Expanded Programme on Immunization (EPI) Programme élargi de vaccination (PEV) Lot Quality Assurance survey to assess immunization coverage Evaluation de la couverture vaccinale par la méthode dite de Lot Quality Assurance (échantillonnage par lots pour l'assurance de la qualité) Burkina Faso. In January 1994, national and provincial Burkina Faso. En janvier 1994, les autorités nationales et provin- public health authorities, in collaboration with WHO, con- ciales de santé publique, en collaboration avec l’OMS, ont mené ducted a field survey to evaluate immunization coverage une étude sur le terrain pour évaluer la couverture vaccinale des for children 12-23 months of age in the city of Bobo enfants de 12 à 23 mois dans la ville de Bobo Dioulasso. L’étude a Dioulasso. The survey was carried out using the method of utilisé la méthode dite de Lot Quality Assurance (LQA) plutôt que Lot Quality Assurance (LQA) rather than the 30-cluster la méthode des 30 grappes plus couramment utilisée par les pro- survey method which has traditionally been used by immu- grammes de vaccination. -

Baseline Study Report
Baseline Study Report MESA II Project - Better Education and Health Agreement: FFE-524-2017/025-00 Final Evaluation Report Coordinated by Project Concern International (PCI) Nicaragua August/Sept. 2017 Submitted to USDA/FAS Project “Mejor Educación y Salud (MESA)” - Nicaragua Agreement: FFE-524-2013-042-00 Submitted to: USDA/FAS Vanessa Castro, José Ramón Laguna, Patricia Callejas with collaboration from Micaela Gómez Managua, December 2017 June 4, 2019 Managua, Nicaragua i Acknowledgements The consultant team appreciates PCI Nicaragua for entrusting Asociación Nicaragua Lee with the completion of this study. In particular, we would like to acknowledge the valuable support provided by María Ángeles Argüello and María Zepeda at PCI Nicaragua-, and by officials from the Ministry of Education (MINED) in Managua and in the departmental delegations of Jinotega and the Southern Caribbean Coast Autonomous Region (RACCS). We also recognize the support given by the officials at the MINED offices in the 11 municipalities participating in the study: Jinotega, La Concordia, San Sebastian de Yali, Santa Maria de Pantasma, Bluefields, Kukra Hill, La Cruz del Río Grande, Laguna de Perlas, Desembocadura Río Grande, El Tortuguero and Corn Island. In particular, we would like to acknowledge the enthusiasm showed by the educational advisors from the aforementioned MINED municipal offices, in the administration of the instruments Our greatest gratitude and consideration to the actors of this study, the fourth-grade students from the elementary schools included in the sample, who agreed and participated with great enthusiasm. We would also like to thank the third-grade teachers who contributed by answering the questionnaire. We should also mention and thank the team of supervisors, applicators and data entry personnel, who put much dedication and effort into the collection and processing of the Early Grade Reading Assessment (EGRA) instruments, the questionnaires, and the school and classroom environment observation sheet. -

Latin American Regional Office
Latin AmericanHONDURAS | 1 Regional Office Newsletter Winter 2020 TABLE OF CONTENTS GUATEMALA 3 First Mesoamerican Meeting on Masculinities 3 Agroecology in Guatemala 4 HONDURAS 5 “I want my life back.” 5 Behind the Scenes 7 NICARAGUA 9 Bertha Madrigal 9 Nohelia Calderon 10 1. Dialogue of Knowledge, Justice and Masculinities Panelists: Jesús Ricardo Sandoval Izaguirre of the Judicial Branch of the Federation Headquarters Monterrey Nuevo León (Mexico), Pedro Rolando Ixchiu García of the Judicial Branch (Guatemala) and Ezequiel González Díaz of the Office of the Attorney General of El Salvador (El Salvador). Photo ©Pepe Orozco 2. Jessy Sandoval, Harlem Padilla, Angélica Rivera, Ivethe Sánchez with the hashtag used in the presentation of the Research on Trafficking, Honduras. Photo ©Ana Judith Aguilar/Calidad de Vida 3. Nohelia Calderon, in her field, which she farms together with her husband. Photo ©Luis Sánchez Corea Graphic Design ©Giulia Vuillermoz/Trócaire. Tegucigalpa, Honduras - January 2020 Latin America Regional Office Newsletter - January 2020 Authors: Ana Maria Alvarez Medrano, Alejandra Guillot Ontanon, Gabriela Flores, Giulia Vuillermoz, Lucia Medina Photographers: Pepe Orozco, Alejandra Guillot Ontanon/Trócaire, Giulia Vuillermoz/Trócaire, Ana Judith Aguilar/ACDV, Luis Sánchez Corea Graphic Design: Giulia Vuillermoz, Trócaire Tegucigalpa, January 2020 GUATEMALA | 3 First experiences, expectations Mesoamerican Meeting on and challenges Masculinities The First Mesoamerican Meeting on Masculinities, Experiences, Expectations and Challenges was held in the departmental capital of the Department of Sololá, Guatemala, from June 26 to 28, 2019, at the facilities of the Centro Cultural Sotz´il Jay (Aldea El Tablón). Women and men (55 men and 52 women), indigenous and mestizo, from ten countries participated: Austria, Bolivia, Canada, Colombia, Costa Rica, El Salvador, Guatemala, Holland, Honduras, Mexico and Nicaragua, including 15 members of Trocaire’s counterparts from Honduras, Nicaragua and Guatemala. -

Annex 8 Nicaragua Country Case Study
NON-EDITED ANNEX 8 NICARAGUA COUNTRY CASE STUDY TABLE OF CONTENTS 1 INTRODUCTION 2 CONTEXT AND SITUATION IN THE COUNTRY REGARDING THE CROSSCUTTING ISSUES 2.1 Political Situation 3 POLICY DIALOGUE: HOW DOES FINLAND EXTEND INFLUENCE AT THE NATIONAL LEVEL IN RELATION TO THE CROSSCUTTING ISSUES 3.1 Bilateral consultations 3.2 Budget Support and Sector Dialogue 3.3 Harmonization and Alignment 3.4 Key Findings – Policy Dialogue 4 MAINSTREAMING IN VARIOUS INTERVENTIONS MODALITIES 4.1 Budget Support Group 4.2 Key Findings – Budget Support 4.3 Sector Support 4.3.1 FONSALUD 4.3.2 PRORURAL 4.4 Key Findings – Sector Support 4.5 Institutionalized Programs 4.5.1 PROGESTION 4.5.2 Key Findings – PROGESTION 4.5.3 FOMEVIDAS 4.5.4 Key Findings – FOMEVIDAS 4.6 Local Cooperation Funds 4.7 Key Findings – Local Cooperation Funds 5 RESPONSABILITIES AND PROCEDURES 5.1 Division of responsibilities between the Embassy and MFA 5.2 Internal Structure for Implementing the Crosscutting issues 5.3 Planning and Management Procedures 5.4 Reporting on Crosscutting Issues 5.5 Key Findings – Responsibilities and Procedures 6 KEY FINDINGS, CONCLUSIONS AND RECOMMENDATIONS 6.1 General Findings 6.2 Findings in the Political Dialogue 6.3 Findings in the Intervention Modalities 6.4 Findings for Responsibilities and Procedures 6.5 Conclusions 6.6 Lessons Learnt 6.7 Recommendations REFERENCES ANNEX 1 PEOPLE INTERVIEWD ACRONYMS AMUNIC Nicaraguan Association of Municipalities BS Budget Support BSWG Budget Support Working Group CED Department Board of Directors CEN National Board of -

World Bank Document
Document of The World Bank Public Disclosure Authorized Report No: ICR2858 IMPLEMENTATION COMPLETION AND RESULTS REPORT (IDA-36650, IDA-3665A, IDA-46800) ON A CREDIT Public Disclosure Authorized IN THE AMOUNT OF SDR 32.50 MILLION (US$42.60 MILLION EQUIVALENT) TO THE REPUBLIC OF NICARAGUA FOR A LAND ADMINISTRATION PROJECT (PRODEP) Public Disclosure Authorized October 20, 2013 Sustainable Development Department Public Disclosure Authorized Central America Country Management Unit Latin America and Caribbean Region CURRENCY EQUIVALENTS (Exchange Rate Effective October 20, 2013) Currency Unit = Cordoba C 24.90 = US$ 1 US$ 1.53 = SDR 1 FISCAL YEAR January 1 - December 31 ABBREVIATIONS AND ACRONYMS ACR Alternative conflict resolution mechanisms ATLMP Agricultural Technology and Land Management Project CIP Comité Interinstitucional del Proyecto (Project Inter-Institutional Committee) CONADETI Comisión Nacional para la Demarcación y Titulación (National Commission for Demarcation and Titling of Indigenous Territories) CPS Country Partnership Strategy CTO Comité Técnico Operativo (Project Operational Technical Committee) CSJ Corte Suprema de Justicia (Nicaraguan Supreme Justice Court) DIRAC Dirección de Resolución Alternativa de Conflictos (Nicaraguan Directorate for Alternative Conflict Resolution under CSJ) DNR Dirección Nacional de Registros (National Directorate of Registries) EA Environmental Assessment EMP Environmental Management Plan FAO Food and Agriculture Organization GPS Global Positioning System ICB International Competitive Bidding -

Nicaragua Expansion and Strengthening Of
PUBLIC SIMULTANEOUS DISCLOSURE DOCUMENT OF THE INTER-AMERICAN DEVELOPMENT BANK NICARAGUA EXPANSION AND STRENGTHENING OF NICARAGUA’S ELECTRICITY TRANSMISSION SYSTEM (NI-L1091) LOAN PROPOSAL This document was prepared by the project team consisting of: Héctor Baldivieso (ENE/CNI), Project Team Leader; Arnaldo Vieira de Carvalho (INE/ENE), Project Team Co-leader; Alberto Levy (INE/ENE); Carlos Trujillo (INE/ENE); Carlos Hinestrosa (INE/ENE); Stephanie Suber (INE/ENE); Juan Carlos Lazo (FMP/CNI); Santiago Castillo (FMP/CNI); María Cristina Landázuri (LEG/SGO); Denis Corrales (VPS/ESG); and Alma Reyna Selva (CID/CNI). This document is being released to the public and distributed to the Bank’s Board of Executive Directors simultaneously. This document has not been approved by the Board. Should the Board approve the document with amendments, a revised version will be made available to the public, thus superseding and replacing the original version. CONTENTS PROJECT SUMMARY I. DESCRIPTION AND RESULTS MONITORING ................................................................ 1 A. Background, problem to be addressed, and rationale ................................... 1 B. Objectives, components, and cost ................................................................ 8 C. Key results indicators ................................................................................. 10 II. FINANCING STRUCTURE AND MAIN RISKS ............................................................... 10 A. Financing instruments ............................................................................... -
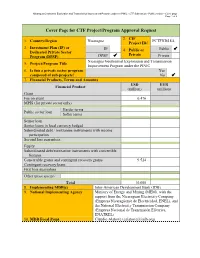
Cover Page for CTF Project/Program Approval Request
Nicaragua Geothermal Exploration and Transmission Improvement Program under the PINIC - CTF Submission - Public version – Cover page Page 1 of 6 Cover Page for CTF Project/Program Approval Request 2. CIF 1. Country/Region Nicaragua PCTFNI618A Project ID# 3. Investment Plan (IP) or IP 4. Public or Public Dedicated Private Sector Private Program (DPSP) DPSP Private Nicaragua Geothermal Exploration and Transmission 5. Project/Program Title Improvement Program under the PINIC 6. Is this a private sector program Yes composed of sub-projects? No 7. Financial Products, Terms and Amounts USD EUR Financial Product (million) (million) Grant Fee on grant 0.476 MPIS (for private sector only) Harder terms Public sector loan Softer terms Senior loan Senior loans in local currency hedged Subordinated debt / mezzanine instruments with income participation Second loss guarantees Equity Subordinated debt/mezzanine instruments with convertible features Convertible grants and contingent recovery grants 9.524 Contingent recovery loans First loss guarantees Other (please specify) Total 10.000 8. Implementing MDB(s) Inter-American Development Bank (IDB) 9. National Implementing Agency Ministry of Energy and Mining (MEM), with the support from the Nicaraguan Electricity Company (Empresa Nicaragüense de Electricidad, ENEL), and the National Electricity Transmission Company (Empresa Nacional de Transmisión Eléctrica, ENATREL) 10. MDB Focal Point Claudio Alatorre ([email protected]) Nicaragua Geothermal Exploration and Transmission Improvement Program under the PINIC - CTF Submission - Public version – Cover page Page 2 of 6 11. Brief Description of Project/Program (including objectives and expected outcomes) In 2015 electricity demand reached 665.4 MW, and it is projected to reach 896 to 1,038 MW by 2026. -

Report (4.723Mb)
Exploration for Bean (Phaseolus) Genetic Resources in Nicaragua December 2007 Technical cooperation between Royal Norwegian Cooperation Utviklingsfondet Centro para la Promoción, la Investigación y el Desarrollo Rural y Social de Nicaragua (CIPRES) Universidad de Costa Rica (UCR) International Center for Tropical Agriculture (CIAT) (photo: Rodolfo Araya; #3202, Dpt. Madríz) Technical Report D.G. Debouck 2 Background and Justification Variability is the engine that drives crop productivity up, and bean does not escape this rule, namely when the bean production in Central America can be heavily affected by other competitive producers with the opening up of borders and with markets becoming global. Understanding the extent of the variability is key to make progress in bean breeding and enhance crop productivity (Singh 1999), and in several market classes of common bean (Voysest 2000) – as the small Central American reds – variability has been demonstrated to be very narrow (Beebe et al. 1995; Sonnante et al. 1994). From here, it is very important to know where variability is, and how variability has evolved through the process of crop domestication and during the recent steps of genetic improvement. In this regard, it is worth understanding the structure of the Mesoamerican races of bean ‘Mesoamerica’ and ‘Guatemala’ (Beebe et al. 2000; Díaz & Blair 2006; Singh et al. 1991), and the variability that may exist in native landraces of Nicaragua (Gómez et al. 2004). By the way, race ‘Mesoamerica’ is the one occupying the largest acreage in the world (Singh 1999), and germplasm of Nicaragua has contributed significantly to the progress of bean breeding (Johnson et al. -

Countries at the Crossroads 2012: Nicaragua
Countries at the Crossroads COUNTRIES AT THE CROSSROADS 2012: NICARAGUA INTRODUCTION Nicaragua’s November 2011 elections marked a major step forward in President Daniel Ortega’s consolidation of power, and a served as a stark demonstration of the authoritarian tendencies he has exhibited since his return to office in early 2007. A onetime guerrilla leader and the head of the Sandinista National Liberation Front (FSLN) during the1979–1990 Sandinista revolution, Ortega has ruled Nicaragua with increasing disrespect for the constitution, electoral integrity, and the rule of law. In order to run again, he engineered a questionable ruling from the Supreme Court of Justice (CSJ) to eviscerate a constitutional ban on successive terms for sitting presidents, as well as a limitation to two total terms of office.1 Ortega then secured 62 percent of the popular vote, although irregularities were widespread enough to cast doubt on the size of his victory margin. The disputed election also gave his party, the FSLN, 63 of 92 seats in the National Assembly, a majority large enough to pass ordinary legislation, change the constitution, and even call a constitutional assembly. Armed with this supermajority, Ortega is now in a position to govern in the temperamental manner of his ideological brethren in the Bolivarian Alliance for the Americas (ALBA) headed by Venezuelan president Hugo Chávez, who has provided him an economic lifeline since 2007. During his first term as president (1984–1990), Ortega presided over the drafting of a constitution in 1987 that reflected the quasi-socialist character of the revolution, which was marked by wealth redistribution and widespread confiscations of private property. -

NICARAGUA Sistema De Información Para El Food Security Situation Seguimiento De La Seguridad Alimentaria Y Nutricional May 2006
Prepared in conjunction with SISSAN NICARAGUA Sistema de Información para el Food Security Situation Seguimiento de la Seguridad Alimentaria y Nutricional May 2006 Alert Levels No Alert Watch Warning Emergency Summary and implications CONTENTS Summary and implications ....................... 1 The beginning of the agricultural cycle is one of the most important activities in the Seasonal calendar ..................................... 1 country in terms of food security. Agriculture provides 21 percent of Nicaragua's gross Current hazards......................................... 1 domestic product, guarantees employment for 38 percent of the economically active Rainy season............................................. 1 population (equivalent to 2 million people) according to the most recent census of the Agricultural production ............................ 2 Nicaraguan Institute of Statistics and Census, and provides more than 40 percent of Prices ........................................................ 3 national exports. The timely beginning of rains is the main factor enabling agricultural Health situation......................................... 3 activities to develop according to expectations, and access to productive means such as seeds and fertilizers is also important. Seasonal calendar Current hazards • High costs of seeds, fertilizers and pesticides, which limit subsistence farmers’ access to these inputs. • Increase in fuel prices, which have generated increases in the prices of transportation, food and other products. • Increase in morbidity due to diarrheic and respiratory diseases that are most prevalent during the beginning of the rainy period. Rainy season Forecasts by the Nicaraguan Institute of Land Studies (INETER) indicate that the rainy period will be established throughout the country in the third dekad of May. An irregular distribution of rains is foreseen in the country, due to variations in both the beginning of the rainy period and the expected precipitation volumes (see Map 1).