Differential Susceptibility to TRAIL of Normal Versus Malignant Human Urothelial Cells
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

TNF Decoy Receptors Encoded by Poxviruses
pathogens Review TNF Decoy Receptors Encoded by Poxviruses Francisco Javier Alvarez-de Miranda † , Isabel Alonso-Sánchez † , Antonio Alcamí and Bruno Hernaez * Centro de Biología Molecular Severo Ochoa, Consejo Superior de Investigaciones Científicas, Campus de Cantoblanco, Universidad Autónoma de Madrid, Nicolás Cabrera 1, 28049 Madrid, Spain; [email protected] (F.J.A.-d.M.); [email protected] (I.A.-S.); [email protected] (A.A.) * Correspondence: [email protected]; Tel.: +34-911-196-4590 † These authors contributed equally. Abstract: Tumour necrosis factor (TNF) is an inflammatory cytokine produced in response to viral infections that promotes the recruitment and activation of leukocytes to sites of infection. This TNF- based host response is essential to limit virus spreading, thus poxviruses have evolutionarily adopted diverse molecular mechanisms to counteract TNF antiviral action. These include the expression of poxvirus-encoded soluble receptors or proteins able to bind and neutralize TNF and other members of the TNF ligand superfamily, acting as decoy receptors. This article reviews in detail the various TNF decoy receptors identified to date in the genomes from different poxvirus species, with a special focus on their impact on poxvirus pathogenesis and their potential use as therapeutic molecules. Keywords: poxvirus; immune evasion; tumour necrosis factor; tumour necrosis factor receptors; lymphotoxin; inflammation; cytokines; secreted decoy receptors; vaccinia virus; ectromelia virus; cowpox virus Citation: Alvarez-de Miranda, F.J.; Alonso-Sánchez, I.; Alcamí, A.; 1. TNF Biology Hernaez, B. TNF Decoy Receptors TNF is a potent pro-inflammatory cytokine with a broad range of biological effects, Encoded by Poxviruses. Pathogens ranging from the activation of inflammatory gene programs to cell differentiation or 2021, 10, 1065. -

TRAIL and Cardiovascular Disease—A Risk Factor Or Risk Marker: a Systematic Review
Journal of Clinical Medicine Review TRAIL and Cardiovascular Disease—A Risk Factor or Risk Marker: A Systematic Review Katarzyna Kakareko 1,* , Alicja Rydzewska-Rosołowska 1 , Edyta Zbroch 2 and Tomasz Hryszko 1 1 2nd Department of Nephrology and Hypertension with Dialysis Unit, Medical University of Białystok, 15-276 Białystok, Poland; [email protected] (A.R.-R.); [email protected] (T.H.) 2 Department of Internal Medicine and Hypertension, Medical University of Białystok, 15-276 Białystok, Poland; [email protected] * Correspondence: [email protected] Abstract: Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is a pro-apoptotic protein showing broad biological functions. Data from animal studies indicate that TRAIL may possibly contribute to the pathophysiology of cardiomyopathy, atherosclerosis, ischemic stroke and abdomi- nal aortic aneurysm. It has been also suggested that TRAIL might be useful in cardiovascular risk stratification. This systematic review aimed to evaluate whether TRAIL is a risk factor or risk marker in cardiovascular diseases (CVDs) focusing on major adverse cardiovascular events. Two databases (PubMed and Cochrane Library) were searched until December 2020 without a year limit in accor- dance to the PRISMA guidelines. A total of 63 eligible original studies were identified and included in our systematic review. Studies suggest an important role of TRAIL in disorders such as heart failure, myocardial infarction, atrial fibrillation, ischemic stroke, peripheral artery disease, and pul- monary and gestational hypertension. Most evidence associates reduced TRAIL levels and increased TRAIL-R2 concentration with all-cause mortality in patients with CVDs. It is, however, unclear Citation: Kakareko, K.; whether low TRAIL levels should be considered as a risk factor rather than a risk marker of CVDs. -
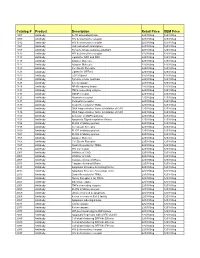
Comprehensive Product List
Catalog # Product Description Retail Price OEM Price 1007 Antibody IL-1R associated kinase 225/100ug 125/100ug 1009 Antibody HIV & chemokine receptor 225/100ug 125/100ug 1012 Antibody HIV & chemokine receptor 225/100ug 125/100ug 1021 Antibody JAK activated transcription 225/100ug 125/100ug 1107 Antibody Tyrosine kinase substrate p62DOK 225/100ug 125/100ug 1112 Antibody HIV & chemokine receptor 225/100ug 125/100ug 1113 Antibody Ligand for DR4 and DR5 225/100ug 125/100ug 1115 Antibody Adapter Molecule 225/100ug 125/100ug 1117 Antibody Adapter Molecule 225/100ug 125/100ug 1120 Antibody Cell Death Receptor 225/100ug 125/100ug 1121 Antibody Ligand for GFRa-2 225/100ug 125/100ug 1123 Antibody CCR3 ligand 225/100ug 125/100ug 1125 Antibody Tyrosine kinase substrate 225/100ug 125/100ug 1128 Antibody A new caspase 225/100ug 125/100ug 1129 Antibody NF-kB inducing kinase 225/100ug 125/100ug 1131 Antibody TNFa converting enzyme 225/100ug 125/100ug 1133 Antibody GDNF receptor 225/100ug 125/100ug 1135 Antibody Neurturin receptor 225/100ug 125/100ug 1137 Antibody Persephin receptor 225/100ug 125/100ug 1139 Antibody Death Receptor for TRAIL 225/100ug 125/100ug 1141 Antibody DNA fragmentation factor & Inhibitor of CAD 225/100ug 125/100ug 1148 Antibody DNA fragmentation factor & Inhibitor of CAD 225/100ug 125/100ug 1150 Antibody Activator of MAPK pathway 225/100ug 125/100ug 1151 Antibody Apoptosis Signal-regulation Kinase 225/100ug 125/100ug 1156 Antibody FLICE inhibitory protein 225/100ug 125/100ug 1158 Antibody Cell Death Receptor 225/100ug 125/100ug 1159 -

Biolegend.Com
Mechanisms of Cell Death TRAIL (TNFSF10) TNF-α Death Receptor 4 (TNFRSF10A/TRAIL-R1) Death Receptor 5 Zombie Dyes (TNFRSF10B/TRAIL-R2) Propidium Iodide (PI) BAT1, TIM-4 TNF RI (TNFRSF1A) 7-Amino-Actinomycin (7-AAD) MER TNF RII (TNFRSF1B) FAS-L GAS6 (TNFSF6/CD178) TRAIL (TNFSF10) Apoptotic Cell Death Domain Zombie Dyes Phosphatidylserine K63 Ubiquitin NH2 Removal ICAM3? ROCK1 NH CD14 2 Eat-Me Signals FAS Death Inducing Cytoskeletal Rearrangement, (TNFRSF6/CD95) Signaling Complex (DISC) TRADD Cytoskeletal Rearrangement, TRADD Decoy Receptor 2 FADD (TNFRSF10D/TRAIL-R4) Actomysin Contraction Engulfment RIP1 TWEAK RIP1 oxLDL (TNFSF12) FADD CIAP1/2 K63 Ubiquitination Blebbing CD36 Death Receptor 3 TWEAK (TNFSF12) PI FADD (TNFRSF25, APO-3) 7-AAD TRAF1 FADD Procaspase 8,10 TRAF 3 Phagocyte FLIP PANX1 Macrophage Monocyte Neutrophil Dendritic Cell Fibroblast Mast Cell Procaspase 8,10 NF-kB TWEAK-R (TNFRSF12A/Fn14) Find-Me Signals Lysophosphocholine C Caspase 8,10 TRAF5 TRAF2 Sphingosine-1-Phosphate G2A? Nucleotides A Decoy TRAIL Receptor R1 (TNFRSF23) Bid Cell Survival ATP, UTP Decoy TRAIL Receptor R2 (TNFRSF22) Sphingosine-1 TRADD Phosphate Receptor Decoy Receptor 1 (TNFRSF10C/TRAIL-R3) Procaspase 3 Proliferation RIP1 G P2y2 t-Bid Bcl-2 T Chemotaxis, Caspase 3 Bcl-2-xL, MCL-1 ? ICAD RIP1 Engulfment Degradation Bax, Bak Oligomerization TRADD Death Receptor 6 Extracellular ATP Bacterial pore-forming toxins TRAIL (TNFSF10) ICAD (TNFRSF21) Monosodium urate crystals Cholesterol crystals Death Receptor DNA Fragmentation Cholera toxin B, Mitochondria -

Cell Structure & Function
Cell Structure & Function Antibodies and Reagents BioLegend is ISO 13485:2016 Certified Toll-Free Tel: (US & Canada): 1.877.BIOLEGEND (246.5343) Tel: 858.768.5800 biolegend.com 02-0012-03 World-Class Quality | Superior Customer Support | Outstanding Value Table of Contents Introduction ....................................................................................................................................................................................3 Cell Biology Antibody Validation .............................................................................................................................................4 Cell Structure/ Organelles ..........................................................................................................................................................8 Cell Development and Differentiation ................................................................................................................................10 Growth Factors and Receptors ...............................................................................................................................................12 Cell Proliferation, Growth, and Viability...............................................................................................................................14 Cell Cycle ........................................................................................................................................................................................16 Cell Signaling ................................................................................................................................................................................18 -

The Concise Guide to Pharmacology 2019/20: Catalytic Receptors
Alexander, S. P. H., Fabbro, D., Kelly, E., Mathie, A., Peters, J. A., Veale, E. L., Armstrong, J. F., Faccenda, E., Harding, S. D., Pawson, A. J., Sharman, J. L., Southan, C., Davies, J. A., & CGTP Collaborators (2019). The Concise Guide to Pharmacology 2019/20: Catalytic receptors. British Journal of Pharmacology, 176(S1), S247- S296. https://doi.org/10.1111/bph.14751 Publisher's PDF, also known as Version of record License (if available): CC BY Link to published version (if available): 10.1111/bph.14751 Link to publication record in Explore Bristol Research PDF-document This is the final published version of the article (version of record). It first appeared online via Wiley at https://bpspubs.onlinelibrary.wiley.com/doi/full/10.1111/bph.14751. Please refer to any applicable terms of use of the publisher. University of Bristol - Explore Bristol Research General rights This document is made available in accordance with publisher policies. Please cite only the published version using the reference above. Full terms of use are available: http://www.bristol.ac.uk/red/research-policy/pure/user-guides/ebr-terms/ S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2019/20: Catalytic receptors. British Journal of Pharmacology (2019) 176, S247–S296 THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Catalytic receptors Stephen PH Alexander1 , Doriano Fabbro2 , Eamonn Kelly3, Alistair Mathie4 ,JohnAPeters5 , Emma L Veale4 , Jane F Armstrong6 , Elena Faccenda6 ,SimonDHarding6 ,AdamJPawson6 , Joanna L Sharman6 , Christopher Southan6 , Jamie A Davies6 -

Cell-Mediated and Cell Membrane-Coated Nanoparticles for Drug Delivery and Cancer Therapy
Yaman et al. Cancer Drug Resist 2020;3:879-911 Cancer DOI: 10.20517/cdr.2020.55 Drug Resistance Review Open Access Cell-mediated and cell membrane-coated nanoparticles for drug delivery and cancer therapy Serkan Yaman1,2,#, Uday Chintapula1,2,#, Edgar Rodriguez1, Harish Ramachandramoorthy1,2, Kytai T. Nguyen1,2 1Department of Bioengineering, University of Texas at Arlington, Arlington, TX 76010, USA. 2Joint Bioengineering Program, University of Texas Southwestern Medical Center, Dallas, TX 75235, USA. #Yaman S and Chintapula U contributed equally to this work. Correspondence to: Dr. Kytai T. Nguyen, Department of Bioengineering, University of Texas at Arlington, 500 UTA Blvd ERB244, Arlington, TX 76010, USA. E-mail: [email protected] How to cite this article: Yaman S, Chintapula U, Rodriguez E, Ramachandramoorthy H, Nguyen KT. Cell-mediated and cell membrane-coated nanoparticles for drug delivery and cancer therapy. Cancer Drug Resist 2020;3:879-911. http://dx.doi.org/10.20517/cdr.2020.55 Received: 24 Jul 2020 First Decision: 25 Aug 2020 Revised: 16 Sep 2020 Accepted: 21 Sep 2020 Available online: 3 Nov 2020 Academic Editor: Vladimir P. Torchilin Copy Editor: Cai-Hong Wang Production Editor: Jing Yu Abstract Nanotechnology-based drug delivery platforms have been developed over the last two decades because of their favorable features in terms of improved drug bioavailability and stability. Despite recent advancement in nanotechnology platforms, this approach still falls short to meet the complexity of biological systems and diseases, such as avoiding systemic side effects, manipulating biological interactions and overcoming drug resistance, which hinders the therapeutic outcomes of the NP-based drug delivery systems. -

A Novel Fully Human Agonistic Single Chain Fragment Variable Antibody Targeting Death Receptor 5 with Potent Antitumor Activity in Vitro and in Vivo
Article A Novel Fully Human Agonistic Single Chain Fragment Variable Antibody Targeting Death Receptor 5 with Potent Antitumor Activity In Vitro and In Vivo Gaoxin Lei, Menglong Xu, Zhipan Xu, Lili Gu, Chenchen Lu, Zhengli Bai, Yue Wang, Yongbo Zhang, Huajing Hu, Yiwei Jiang, Wenfeng Zhao and Shuhua Tan * State Key Laboratory of Natural Medicines, School of Life Science and Technology, China Pharmaceutical University, Nanjing 210009, China; [email protected] (G.L.); [email protected] (M.X.); [email protected] (Z.X.); [email protected] (L.G.); [email protected] (C.L.); [email protected] (Z.B.); [email protected] (Y.W.); [email protected] (Y.Z.); [email protected] (H.H.); [email protected] (Y.J.); [email protected] (W.Z.) * Correspondence: [email protected]; Tel.: +86-25-8327-1012 Received: 27 July 2017; Accepted: 17 September 2017; Published: 27 September 2017 Abstract: Agonistic antibodies, which bind specifically to death receptor 5 (DR5), can trigger apoptosis in tumor cells through the extrinsic pathway. In this present study, we describe the use of a phage display to isolate a novel fully human agonistic single chain fragment variable (scFv) antibody, which targets DR5. After five rounds of panning a large (1.2 × 108 clones) phage display library on DR5, a total of over 4000 scFv clones were screened by the phage ELISA. After screening for agonism in a cell-viability assay in vitro, a novel DR5-specific scFv antibody TR2-3 was isolated, which inhibited COLO205 and MDA-MB-231 tumor cell growth without any cross-linking agents. -

TRAIL-Decoy Receptor-1 Disappears in Granulosa Cells of Atretic Follicles in Porcine Ovaries
Journal of Reproduction and Development, Vol. 48, No. 2, 2002 —Original— TRAIL-Decoy Receptor-1 Disappears in Granulosa Cells of Atretic Follicles in Porcine Ovaries Satoko WADA1), Noboru MANABE1), Naoko INOUE1), Mizuho NAKAYAMA1), Toshikatsu MATSUI1) and Hajime MIYAMOTO1) 1) Unit of Anatomy and Cell Biology, Department of Animal Sciences, Kyoto University, Kyoto 606-8502, Japan Abstract. To reveal the specific regulatory molecules that control granulosa cell apoptosis during follicular atresia, we immunohistochemically examined the localization of tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) and its receptors in porcine ovaries. A marked reduction in the expression of decoy receptor-1 (DcR1), which has high affinity for TRAIL, was demonstrated in granulosa cells of atretic follicles, but no marked differences were seen in expression of TRAIL or other TRAIL-receptors (death receptor-4 or death receptor-5) in granulosa cells between healthy and atretic follicles. No positive staining for DcR2 was seen. We presum that TRAIL and its receptors are involved in induction of apoptosis in granulosa cells during atresia, and that DcR1 plays an inhibitory role in granulosa cell apoptosis. Key words: Apoptosis, TRAIL-decoy receptor, Follicular atresia, Granulosa cell, Pig (J. Reprod. Dev. 48: 167–173, 2002) n mammalian ovaries, more than 99% of the [15–19]. However, it has not been determined follicles undergo a degenerative change known whether the FasL-Fas system mediates apoptosis in as atresia at varying stages of follicle development pig ovaries. Previously, we revealed species- [1, 2]. A number of studies of follicular atresia have specific differences in the apoptotic process in revealed the morphological and biochemical granulosa cells and indicated that local mechanisms characteristics of atretic follicles [3, 4]. -
Characterization of Chicken TNFR Superfamily Decoy Receptors, Dcr3 and Osteoprotegerinq
BBRC Biochemical and Biophysical Research Communications 307 (2003) 956–961 www.elsevier.com/locate/ybbrc Characterization of chicken TNFR superfamily decoy receptors, DcR3 and osteoprotegerinq Jamie T. Bridgham and Alan L. Johnson* Department of Biological Sciences and the Walther Cancer Research Center, The University of Notre Dame, P.O. Box369, Notre Dame, IN 46556, USA Received 31 May 2003 Abstract Tumor necrosis factor (TNF) family ligands bind to death domain-containing TNF receptors (death receptors), which can subsequently activate intracellular signaling pathways to initiate caspase activity and apoptotic cell death. Decoy receptors, without intracellular death domains, have been reported to prevent cytotoxic effects by binding to and sequestering such ligands, or by interfering with death receptor trimerization. The chicken death receptors, Fas, TNFR1, DR6, and TVB, are constitutively ex- pressed in a relatively wide variety of hen tissues. In this study, two chicken receptors with sequence homology to the mammalian decoys, DcR3 and osteoprotegerin, were identified and their pattern of expression was characterized. Unlike the previously iden- tified chicken death receptors, the newly characterized decoy receptors show comparatively limited expression among tissues, suggesting a tissue-specific function. Finally, characterization of these chicken receptors further contributes to understanding the evolutionary divergence of TNFR superfamily members among vertebrate species. Ó 2003 Elsevier Inc. All rights reserved. Keywords: Tumor necrosis factor receptor superfamily; TNF ligands; Death receptor; Decoy receptor; Ovary; Chicken; Apoptosis Death receptors (DRs) are characterized by the pres- Alternatively, many of the death receptors are capa- ence of a variable number of extracellular cysteine-rich ble of signaling through intracellular pathways that ac- motifs that comprise the ligand binding domain, a single- tually promote cell survival. -
University of Groningen Probing the Ligand Receptor
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by University of Groningen University of Groningen Probing the ligand receptor interface of TNF ligand family members RANKL and TRAIL Wang, Yizhou DOI: 10.33612/diss.127959201 IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it. Please check the document version below. Document Version Publisher's PDF, also known as Version of record Publication date: 2020 Link to publication in University of Groningen/UMCG research database Citation for published version (APA): Wang, Y. (2020). Probing the ligand receptor interface of TNF ligand family members RANKL and TRAIL. University of Groningen. https://doi.org/10.33612/diss.127959201 Copyright Other than for strictly personal use, it is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), unless the work is under an open content license (like Creative Commons). Take-down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. Downloaded from the University of Groningen/UMCG research database (Pure): http://www.rug.nl/research/portal. For technical reasons the number of authors shown on this cover page is limited to 10 maximum. Download date: 26-12-2020 Chapter 1 General introduction and scope of the thesis Chapter 1 TNF superfamily The discovery of tumor necrosis factor (TNF) superfamily dates back to the middle of the nineteenth century when O‘Malley found a tumor-necrotizing factor in the serum that could mediate tumor regression effects [1]. -
Human CD263 / TRAIL-R3 ELISA Kit (ARG81333)
Product datasheet [email protected] ARG81333 Package: 96 wells Human CD263 / TRAIL-R3 ELISA Kit Store at: 4°C Summary Product Description ARG81333 Human CD263 / TRAIL-R3 ELISA Kit is an Enzyme Immunoassay kit for the quantification of Human CD263 / TRAIL-R3 in serum, plasma and cell culture supernatant. Tested Reactivity Hu Tested Application ELISA Specificity Specific for Human CD263 / TRAIL-R3. No cross reactivity was observed with TRAIL, CD117, IL-6R, IL-2R, CD116, TRAIL R1, TRAIL R2, TRAIL R4, CD178, and Granzyme B. Target Name CD263 / TRAIL R3 Conjugation HRP Conjugation Note Substrate: TMB and read at 450 nm. Sensitivity 147 pg/ml Sample Type Serum, plasma and cell culture supernatants. Standard Range 312.5 - 10000 pg/ml Sample Volume 100 µl Alternate Names Lymphocyte inhibitor of TRAIL; Antagonist decoy receptor for TRAIL/Apo-2L; TNF-related apoptosis- inducing ligand receptor 3; DCR1; TRID; CD antigen CD263; Tumor necrosis factor receptor superfamily member 10C; CD263; Decoy TRAIL receptor without death domain; LIT; Decoy receptor 1; DcR1; DCR1-TNFR; TRAIL-R3; TRAIL receptor 3; TRAILR3; TRAIL receptor without an intracellular domain Application Instructions Assay Time ~ 4 hours Properties Form 96 well Storage instruction Store the kit at 2-8°C. Keep microplate wells sealed in a dry bag with desiccants. Do not expose test reagents to heat, sun or strong light during storage and usage. Please refer to the product user manual for detail temperatures of the components. Note For laboratory research only, not for drug, diagnostic or other use. Bioinformation Gene Symbol TNFRSF10C Gene Full Name tumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain Background The protein encoded by this gene is a member of the TNF-receptor superfamily.