The Concise Guide to Pharmacology 2019/20: Catalytic Receptors
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

TNF Decoy Receptors Encoded by Poxviruses
pathogens Review TNF Decoy Receptors Encoded by Poxviruses Francisco Javier Alvarez-de Miranda † , Isabel Alonso-Sánchez † , Antonio Alcamí and Bruno Hernaez * Centro de Biología Molecular Severo Ochoa, Consejo Superior de Investigaciones Científicas, Campus de Cantoblanco, Universidad Autónoma de Madrid, Nicolás Cabrera 1, 28049 Madrid, Spain; [email protected] (F.J.A.-d.M.); [email protected] (I.A.-S.); [email protected] (A.A.) * Correspondence: [email protected]; Tel.: +34-911-196-4590 † These authors contributed equally. Abstract: Tumour necrosis factor (TNF) is an inflammatory cytokine produced in response to viral infections that promotes the recruitment and activation of leukocytes to sites of infection. This TNF- based host response is essential to limit virus spreading, thus poxviruses have evolutionarily adopted diverse molecular mechanisms to counteract TNF antiviral action. These include the expression of poxvirus-encoded soluble receptors or proteins able to bind and neutralize TNF and other members of the TNF ligand superfamily, acting as decoy receptors. This article reviews in detail the various TNF decoy receptors identified to date in the genomes from different poxvirus species, with a special focus on their impact on poxvirus pathogenesis and their potential use as therapeutic molecules. Keywords: poxvirus; immune evasion; tumour necrosis factor; tumour necrosis factor receptors; lymphotoxin; inflammation; cytokines; secreted decoy receptors; vaccinia virus; ectromelia virus; cowpox virus Citation: Alvarez-de Miranda, F.J.; Alonso-Sánchez, I.; Alcamí, A.; 1. TNF Biology Hernaez, B. TNF Decoy Receptors TNF is a potent pro-inflammatory cytokine with a broad range of biological effects, Encoded by Poxviruses. Pathogens ranging from the activation of inflammatory gene programs to cell differentiation or 2021, 10, 1065. -

TRAIL and Cardiovascular Disease—A Risk Factor Or Risk Marker: a Systematic Review
Journal of Clinical Medicine Review TRAIL and Cardiovascular Disease—A Risk Factor or Risk Marker: A Systematic Review Katarzyna Kakareko 1,* , Alicja Rydzewska-Rosołowska 1 , Edyta Zbroch 2 and Tomasz Hryszko 1 1 2nd Department of Nephrology and Hypertension with Dialysis Unit, Medical University of Białystok, 15-276 Białystok, Poland; [email protected] (A.R.-R.); [email protected] (T.H.) 2 Department of Internal Medicine and Hypertension, Medical University of Białystok, 15-276 Białystok, Poland; [email protected] * Correspondence: [email protected] Abstract: Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is a pro-apoptotic protein showing broad biological functions. Data from animal studies indicate that TRAIL may possibly contribute to the pathophysiology of cardiomyopathy, atherosclerosis, ischemic stroke and abdomi- nal aortic aneurysm. It has been also suggested that TRAIL might be useful in cardiovascular risk stratification. This systematic review aimed to evaluate whether TRAIL is a risk factor or risk marker in cardiovascular diseases (CVDs) focusing on major adverse cardiovascular events. Two databases (PubMed and Cochrane Library) were searched until December 2020 without a year limit in accor- dance to the PRISMA guidelines. A total of 63 eligible original studies were identified and included in our systematic review. Studies suggest an important role of TRAIL in disorders such as heart failure, myocardial infarction, atrial fibrillation, ischemic stroke, peripheral artery disease, and pul- monary and gestational hypertension. Most evidence associates reduced TRAIL levels and increased TRAIL-R2 concentration with all-cause mortality in patients with CVDs. It is, however, unclear Citation: Kakareko, K.; whether low TRAIL levels should be considered as a risk factor rather than a risk marker of CVDs. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Integrin Beta 7 Polyclonal Antibody Purified Rabbit Polyclonal Antibody (Pab) Catalog # AP54281
10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 Integrin beta 7 Polyclonal Antibody Purified Rabbit Polyclonal Antibody (Pab) Catalog # AP54281 Specification Integrin beta 7 Polyclonal Antibody - Product Information Application IHC-P Primary Accession P26011 Reactivity Rat Host Rabbit Clonality Polyclonal Calculated MW 87411 Integrin beta 7 Polyclonal Antibody - Additional Information Gene ID 16421 Tissue/cell: human glioma tissue; 4% Paraformaldehyde-fixed and Other Names paraffin-embedded; Integrin beta-7, Integrin beta-P, M290 IEL Antigen retrieval: citrate buffer ( 0.01M, pH antigen, Itgb7 6.0 ), Boiling bathing for 15min; Block Format endogenous peroxidase by 3% Hydrogen 0.01M TBS(pH7.4) with 1% BSA, 0.09% peroxide for 30min; Blocking buffer (normal (W/V) sodium azide and 50% Glyce goat serum,C-0005) at 37℃ for 20 min; Incubation: Anti-Integrin beta 7 Polyclonal Storage Antibody, Unconjugated(bs-1051R) 1:200, Store at -20 ℃ for one year. Avoid repeated overnight at 4°C, followed by conjugation to freeze/thaw cycles. When reconstituted in the secondary antibody(SP-0023) and sterile pH 7.4 0.01M PBS or diluent of DAB(C-0010) staining antibody the antibody is stable for at least two weeks at 2-4 ℃. Integrin beta 7 Polyclonal Antibody - Protein Information Name Itgb7 Function Integrin alpha-4/beta-7 (Peyer patches-specific homing receptor LPAM-1) is involved in adhesive interactions of leukocytes. It is a receptor for fibronectin and recognizes one or more domains within the alternatively spliced CS-1 region of fibronectin. Integrin alpha- 4/beta-7 is also a receptor for MADCAM1 and VCAM1. -
![Cd49d Antibody [R1-2] Cat](https://docslib.b-cdn.net/cover/1976/cd49d-antibody-r1-2-cat-571976.webp)
Cd49d Antibody [R1-2] Cat
CD49d Antibody [R1-2] Cat. No.: 76-809 CD49d Antibody [R1-2] Specifications HOST SPECIES: Rat SPECIES REACTIVITY: Mouse TESTED APPLICATIONS: Flow, Func The R1-2 monoclonal antibody specifically reacts with mouse CD49d (integrin alpha 4), SPECIFICITY: which forms the VLA-4 complex with CD29 (integrin beta 1). Properties The monoclonal antibody was purified utilizing affinity chromatography. The endotoxin PURIFICATION: level is determined by LAL test to be less than 0.01 EU/µg of the protein. CLONALITY: Monoclonal ISOTYPE: Rat IgG2b, kappa CONJUGATE: Unconjugated PHYSICAL STATE: liquid BUFFER: Phosphate-buffered aqueous solution, ph7.2. CONCENTRATION: batch dependent STORAGE CONDITIONS: The product should be stored undiluted at 4˚C . Do not freeze. September 27, 2021 1 https://www.prosci-inc.com/cd49d-antibody-r1-2-76-809.html Additional Info OFFICIAL SYMBOL: Itga4 ALTERNATE NAMES: CD49D, Itga4B, Itga4 GENE ID: 16401 USER NOTE: Optimal dilutions for each application to be determined by the researcher. Background and References The R1-2 monoclonal antibody specifically reacts with mouse CD49d (integrin alpha 4), which forms the VLA-4 complex with CD29 (integrin beta 1). VLA-4 is expressed on BACKGROUND: thymocytes, monocytes, and a subset of peripheral lymphocytes. It is the receptor for fibronectin and CD106 (VCAM-1). CD49d also forms a receptor for mucosal vascular addressin (MAdCAM-1) with integrin beta 7. 1) Deckert, M., Kubar, J., Zoccola, D., Bernard‐Pomier, G., Angelisova, P., Horejsi, V., REFERENCES: Bernard, A. (1992). CD59 molecule: a second ligand for CD2 in T cell adhesion.European journal of immunology,22(11), 2943-2947. 2) Korty, P. -

Silencing of ITGB6 Inhibits the Progression of Cervical Carcinoma Via Regulating JAK/STAT3 Signaling Pathway
803 Original Article Page 1 of 12 Silencing of ITGB6 inhibits the progression of cervical carcinoma via regulating JAK/STAT3 signaling pathway Xiaoxia Zheng, Yanan Zhu, Xiaoping Wang, Yanmei Hou, Yingji Fang Jinan Maternity and Child Care Hospital Affiliated to Shandong First Medical University/Jinan Maternity and Child Care Hospital Affiliated, Jinan, China Contributions: (I) Conception and design: Y Fang; (II) Administrative support: Y Hou; (III) Provision of study materials or patients: X Zheng, Y Zhu, X Wang; (IV) Collection and assembly of data: X Zheng, Y Zhu, X Wang; (V) Data analysis and interpretation: X Zheng, Y Zhu, X Wang; (VI) Manuscript writing: All authors; (VII) Final approval of manuscript: All authors. Correspondence to: Yingji Fang. Jinan Maternity and Child Care Hospital Affiliated to Shandong First Medical University/Jinan Maternity and Child Care Hospital Affiliated, No. 2 Jianguo Xiaojingsan Road, Jinan 250001, China. Email:[email protected] . Background: Integrin β6 (ITGB6), a key submonomer of integrin αvβ6, plays an important role in epithelial-to-mesenchymal transition (EMT), wound healing, epithelial-derived tumor growth, fibrosis, and epithelial repair. However, the role of ITGB6 in cervical carcinoma (CC) remains elusive. Methods: The expression levels of ITGB6 in CC tissues and cell lines were determined using quantitative real-time polymerase chain reaction (qRT-PCR). The cell viability, proliferation, apoptosis, migration, and invasion were evaluated by Cell Counting Kit-8 (CCK-8), colony-forming, flow cytometry, and Transwell assay, respectively. The expression of related proteins, including EMT markers and the Janus kinase/signal transducer and activator of transcription (JAK/STAT3) signaling markers, were detected using western blotting. -
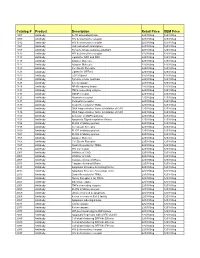
Comprehensive Product List
Catalog # Product Description Retail Price OEM Price 1007 Antibody IL-1R associated kinase 225/100ug 125/100ug 1009 Antibody HIV & chemokine receptor 225/100ug 125/100ug 1012 Antibody HIV & chemokine receptor 225/100ug 125/100ug 1021 Antibody JAK activated transcription 225/100ug 125/100ug 1107 Antibody Tyrosine kinase substrate p62DOK 225/100ug 125/100ug 1112 Antibody HIV & chemokine receptor 225/100ug 125/100ug 1113 Antibody Ligand for DR4 and DR5 225/100ug 125/100ug 1115 Antibody Adapter Molecule 225/100ug 125/100ug 1117 Antibody Adapter Molecule 225/100ug 125/100ug 1120 Antibody Cell Death Receptor 225/100ug 125/100ug 1121 Antibody Ligand for GFRa-2 225/100ug 125/100ug 1123 Antibody CCR3 ligand 225/100ug 125/100ug 1125 Antibody Tyrosine kinase substrate 225/100ug 125/100ug 1128 Antibody A new caspase 225/100ug 125/100ug 1129 Antibody NF-kB inducing kinase 225/100ug 125/100ug 1131 Antibody TNFa converting enzyme 225/100ug 125/100ug 1133 Antibody GDNF receptor 225/100ug 125/100ug 1135 Antibody Neurturin receptor 225/100ug 125/100ug 1137 Antibody Persephin receptor 225/100ug 125/100ug 1139 Antibody Death Receptor for TRAIL 225/100ug 125/100ug 1141 Antibody DNA fragmentation factor & Inhibitor of CAD 225/100ug 125/100ug 1148 Antibody DNA fragmentation factor & Inhibitor of CAD 225/100ug 125/100ug 1150 Antibody Activator of MAPK pathway 225/100ug 125/100ug 1151 Antibody Apoptosis Signal-regulation Kinase 225/100ug 125/100ug 1156 Antibody FLICE inhibitory protein 225/100ug 125/100ug 1158 Antibody Cell Death Receptor 225/100ug 125/100ug 1159 -

Integrin Β6 Serves As an Immunohistochemical Marker For
www.nature.com/scientificreports OPEN Integrin β6 serves as an immunohistochemical marker for lymph node metastasis and Received: 21 April 2016 Accepted: 29 June 2016 promotes cell invasiveness in Published: 21 July 2016 cholangiocarcinoma Zequn Li1,2, Siddhartha Biswas1,2, Benjia Liang1,2, Xueqing Zou1,2, Liqun Shan1,2, Yang Li1,2, Ruliang Fang1 & Jun Niu1 Cholangiocarcinoma is a devastating malignancy that is notoriously difficult to diagnose and is associated with a high mortality. Despite extensive efforts to improve the diagnosis and treatment of this neoplasm, limited progress has been made. Integrin β6 is a subtype of integrin that is expressed exclusively on the surfaces of epithelial cells and is associated with a variety of tumors. In the present study, we investigated the expression and roles of integrin β6 in cholangiocarcinoma. β6 upregulation in cholangiocarcinoma was correlated with lymph node metastasis and distant metastasis. Moreover, integrin β6 was identified as a biomarker for the diagnosis of cholangiocarcinoma and an indicator of lymph node metastasis. Integrin β6 significantly promoted the proliferation, migration and invasion of cholangiocarcinoma cells. Furthermore, integrin β6 increased Rac1-GTPase, resulting in the upregulation of metalloproteinase-9 (MMP9) and F-actin polymerization. Taken together, our results indicate that integrin β6 promotes tumor invasiveness in a Rac1-dependent manner and is a potential biomarker for tumor metastasis. Integrin β6 may help to improve the diagnostic accuracy, and targeting β6 may be a novel strategy for the treatment of cholangiocarcinoma. Cholangiocarcinoma is neoplasm originating from the ductular epithelium of the biliary tree, either within the liver (intrahepatic cholangiocarcinoma) or more commonly from the extrahepatic bile ducts (extrahepatic cholan- giocarcinoma). -

Supp Table 6.Pdf
Supplementary Table 6. Processes associated to the 2037 SCL candidate target genes ID Symbol Entrez Gene Name Process NM_178114 AMIGO2 adhesion molecule with Ig-like domain 2 adhesion NM_033474 ARVCF armadillo repeat gene deletes in velocardiofacial syndrome adhesion NM_027060 BTBD9 BTB (POZ) domain containing 9 adhesion NM_001039149 CD226 CD226 molecule adhesion NM_010581 CD47 CD47 molecule adhesion NM_023370 CDH23 cadherin-like 23 adhesion NM_207298 CERCAM cerebral endothelial cell adhesion molecule adhesion NM_021719 CLDN15 claudin 15 adhesion NM_009902 CLDN3 claudin 3 adhesion NM_008779 CNTN3 contactin 3 (plasmacytoma associated) adhesion NM_015734 COL5A1 collagen, type V, alpha 1 adhesion NM_007803 CTTN cortactin adhesion NM_009142 CX3CL1 chemokine (C-X3-C motif) ligand 1 adhesion NM_031174 DSCAM Down syndrome cell adhesion molecule adhesion NM_145158 EMILIN2 elastin microfibril interfacer 2 adhesion NM_001081286 FAT1 FAT tumor suppressor homolog 1 (Drosophila) adhesion NM_001080814 FAT3 FAT tumor suppressor homolog 3 (Drosophila) adhesion NM_153795 FERMT3 fermitin family homolog 3 (Drosophila) adhesion NM_010494 ICAM2 intercellular adhesion molecule 2 adhesion NM_023892 ICAM4 (includes EG:3386) intercellular adhesion molecule 4 (Landsteiner-Wiener blood group)adhesion NM_001001979 MEGF10 multiple EGF-like-domains 10 adhesion NM_172522 MEGF11 multiple EGF-like-domains 11 adhesion NM_010739 MUC13 mucin 13, cell surface associated adhesion NM_013610 NINJ1 ninjurin 1 adhesion NM_016718 NINJ2 ninjurin 2 adhesion NM_172932 NLGN3 neuroligin -

Biolegend.Com
Mechanisms of Cell Death TRAIL (TNFSF10) TNF-α Death Receptor 4 (TNFRSF10A/TRAIL-R1) Death Receptor 5 Zombie Dyes (TNFRSF10B/TRAIL-R2) Propidium Iodide (PI) BAT1, TIM-4 TNF RI (TNFRSF1A) 7-Amino-Actinomycin (7-AAD) MER TNF RII (TNFRSF1B) FAS-L GAS6 (TNFSF6/CD178) TRAIL (TNFSF10) Apoptotic Cell Death Domain Zombie Dyes Phosphatidylserine K63 Ubiquitin NH2 Removal ICAM3? ROCK1 NH CD14 2 Eat-Me Signals FAS Death Inducing Cytoskeletal Rearrangement, (TNFRSF6/CD95) Signaling Complex (DISC) TRADD Cytoskeletal Rearrangement, TRADD Decoy Receptor 2 FADD (TNFRSF10D/TRAIL-R4) Actomysin Contraction Engulfment RIP1 TWEAK RIP1 oxLDL (TNFSF12) FADD CIAP1/2 K63 Ubiquitination Blebbing CD36 Death Receptor 3 TWEAK (TNFSF12) PI FADD (TNFRSF25, APO-3) 7-AAD TRAF1 FADD Procaspase 8,10 TRAF 3 Phagocyte FLIP PANX1 Macrophage Monocyte Neutrophil Dendritic Cell Fibroblast Mast Cell Procaspase 8,10 NF-kB TWEAK-R (TNFRSF12A/Fn14) Find-Me Signals Lysophosphocholine C Caspase 8,10 TRAF5 TRAF2 Sphingosine-1-Phosphate G2A? Nucleotides A Decoy TRAIL Receptor R1 (TNFRSF23) Bid Cell Survival ATP, UTP Decoy TRAIL Receptor R2 (TNFRSF22) Sphingosine-1 TRADD Phosphate Receptor Decoy Receptor 1 (TNFRSF10C/TRAIL-R3) Procaspase 3 Proliferation RIP1 G P2y2 t-Bid Bcl-2 T Chemotaxis, Caspase 3 Bcl-2-xL, MCL-1 ? ICAD RIP1 Engulfment Degradation Bax, Bak Oligomerization TRADD Death Receptor 6 Extracellular ATP Bacterial pore-forming toxins TRAIL (TNFSF10) ICAD (TNFRSF21) Monosodium urate crystals Cholesterol crystals Death Receptor DNA Fragmentation Cholera toxin B, Mitochondria -

Cell Adhesion Molecules in Normal Skin and Melanoma
biomolecules Review Cell Adhesion Molecules in Normal Skin and Melanoma Cian D’Arcy and Christina Kiel * Systems Biology Ireland & UCD Charles Institute of Dermatology, School of Medicine, University College Dublin, D04 V1W8 Dublin, Ireland; [email protected] * Correspondence: [email protected]; Tel.: +353-1-716-6344 Abstract: Cell adhesion molecules (CAMs) of the cadherin, integrin, immunoglobulin, and selectin protein families are indispensable for the formation and maintenance of multicellular tissues, espe- cially epithelia. In the epidermis, they are involved in cell–cell contacts and in cellular interactions with the extracellular matrix (ECM), thereby contributing to the structural integrity and barrier for- mation of the skin. Bulk and single cell RNA sequencing data show that >170 CAMs are expressed in the healthy human skin, with high expression levels in melanocytes, keratinocytes, endothelial, and smooth muscle cells. Alterations in expression levels of CAMs are involved in melanoma propagation, interaction with the microenvironment, and metastasis. Recent mechanistic analyses together with protein and gene expression data provide a better picture of the role of CAMs in the context of skin physiology and melanoma. Here, we review progress in the field and discuss molecular mechanisms in light of gene expression profiles, including recent single cell RNA expression information. We highlight key adhesion molecules in melanoma, which can guide the identification of pathways and Citation: D’Arcy, C.; Kiel, C. Cell strategies for novel anti-melanoma therapies. Adhesion Molecules in Normal Skin and Melanoma. Biomolecules 2021, 11, Keywords: cadherins; GTEx consortium; Human Protein Atlas; integrins; melanocytes; single cell 1213. https://doi.org/10.3390/ RNA sequencing; selectins; tumour microenvironment biom11081213 Academic Editor: Sang-Han Lee 1. -

Integrin Beta 7 Antibody (F44498)
Integrin beta 7 Antibody (F44498) Catalog No. Formulation Size F44498-0.4ML In 1X PBS, pH 7.4, with 0.09% sodium azide 0.4 ml F44498-0.08ML In 1X PBS, pH 7.4, with 0.09% sodium azide 0.08 ml Bulk quote request Availability 1-3 business days Species Reactivity Human Format Antigen affinity purified Clonality Polyclonal (rabbit origin) Isotype Rabbit Ig Purity Antigen affinity UniProt P26010 Applications Western blot : 1:1000 Limitations This Integrin beta 7 antibody is available for research use only. Integrin beta 7 antibody western blot analysis in WiDr lysate. Description Integrin alpha-4/beta-7 (Peyer patches-specific homing receptor LPAM-1) is an adhesion molecule that mediates lymphocyte migration and homing to gut-associated lymphoid tissue (GALT). Integrin alpha-4/beta-7 interacts with the cell surface adhesion molecules MADCAM1 which is normally expressed by the vascular endothelium of the gastrointestinal tract. Interacts also with VCAM1 and fibronectin, an extracellular matrix component. It recognizes one or more domains within the alternatively spliced CS-1 region of fibronectin. Interactions involves the tripeptide L-D-T in MADCAM1, and L-D-V in fibronectin. Binds to HIV-1 gp120, thereby allowing the virus to enter GALT, which is thought to be the major trigger of AIDS disease. Interaction would involve a tripeptide L-D-I in HIV-1 gp120. Integrin alpha-E/beta-7 (HML-1) is a receptor for E-cadherin. Application Notes Titration of the Integrin beta 7 antibody may be required due to differences in protocols and secondary/substrate sensitivity.