A Phylogenetic Analysis of Antrodia S.L. Based on Nrdna ITS Sequences, with Emphasis on Rhizomorphic European Species
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
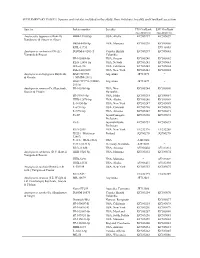
Suppl Table I
SUPPLEMENTARY TABLE I. Species and isolates included in the study, their vouchers, locality and GenBank accession Species Isolate number Locality ITS GenBank LSU GenBank accession no. accession no. Amylocystis lapponica (Romell) HHB-13400-Sp USA, Alaska KC585237 KC585059 Bondartsev & Singer ex Singer OKM-4418-Sp USA, Montana KC585238 KC585060 KHL-11755 − EU118603 Amyloporia carbonica (Overh.) DAOM-F-8281-T Canada, British KC585239 KC585061 Vampola & Pouzar Columbia FP-105585-Sp USA, Oregon KC585240 KC585062 RLG-12496-Sp USA, Nevada KC585241 KC585063 Wilcox-96 USA, California KC585242 KC585064 Zabel-40-GLN USA, New York KC585243 KC585065 Amyloporia nothofaginea Rajchenb. BAFC519794 Argentina JF713078 − & Gorjón (=MMBP-2011) BAFC519796 (=MBP- Argentina JF713079 − 2011a) Amyloporia sinuosa (Fr.) Rajchenb., FP-105386-Sp USA, New KC585244 KC585066 Gorjón & Pildain Hampshire FP-94464-Sp USA, Idaho KC585245 KC585067 HHB-12878-Sp USA, Alaska KC585246 KC585068 L-14130-Sp USA, New York KC585247 KC585069 L-6192-Sp USA, Colorado KC585248 KC585070 L-9792-Sp USA, Arizona KC585249 KC585071 Pa-3C JapanYamagata KC585250 KC585072 Prefecture Pa-3e JapanShizuoka KC585251 KC585073 Prefecture RLG-2538 USA, New York EU232196 EU232288 X725 (=Miettinen- Finland JQ700270 JQ700270 12407) P-115 (=RLG-2538) USA AJ416068 − P-211 (G-214) Germany, Karlsrube AJ345011 − RLG-1182R USA, Arizona AY966450 AY333831 Amyloporia sitchensis (D.V. Baxter) HHB-5320-Sp USA, Montana KC585252 KC585074 Vampola & Pouzar HHB-5298 USA, Montana − AY333829 HHB-12513 USA, Alaska AY966451 AY333830 -

Short-Read Sequencing for Genomic Analysis of the Brown Rot Fungus Fibroporia Radiculosa
Short-Read Sequencing for Genomic Analysis of the Brown Rot Fungus Fibroporia radiculosa Juliet D. Tang,a Andy D. Perkins,b Tad S. Sonstegard,c Steven G. Schroeder,c Shane C. Burgess,d and Susan V. Diehla Forest Products, Mississippi State University, Mississippi State, Mississippi, USAa; Computer Science and Engineering, Mississippi State University, Mississippi State, Mississippi, USAb; USDA ARS Bovine Functional Genomics Laboratory, Beltsville, Maryland, USAc; and Institute for Genomics, Biocomputing, and Biotechnology, Mississippi State University, Mississippi State, Mississippi, USAd The feasibility of short-read sequencing for genomic analysis was demonstrated for Fibroporia radiculosa, a copper-tolerant fungus that causes brown rot decay of wood. The effect of read quality on genomic assembly was assessed by filtering Illumina GAIIx reads from a single run of a paired-end library (75-nucleotide read length and 300-bp fragment size) at three different stringency levels and then assembling each data set with Velvet. A simple approach was devised to determine which filter strin- gency was “best.” Venn diagrams identified the regions containing reads that were used in an assembly but were of a low-enough quality to be removed by a filter. By plotting base quality histograms of reads in this region, we judged whether a filter was too stringent or not stringent enough. Our best assembly had a genome size of 33.6 Mb, an N50 of 65.8 kb for a k-mer of 51, and a maximum contig length of 347 kb. Using GeneMark, 9,262 genes were predicted. TargetP and SignalP analyses showed that among the 1,213 genes with secreted products, 986 had motifs for signal peptides and 227 had motifs for signal anchors. -

A New Species of Antrodia (Basidiomycota, Polypores) from China
Mycosphere 8(7): 878–885 (2017) www.mycosphere.org ISSN 2077 7019 Article Doi 10.5943/mycosphere/8/7/4 Copyright © Guizhou Academy of Agricultural Sciences A new species of Antrodia (Basidiomycota, Polypores) from China Chen YY, Wu F* Institute of Microbiology, Beijing Forestry University, Beijing 100083, China Chen YY, Wu F 2017 –A new species of Antrodia (Basidiomycota, Polypores) from China. Mycosphere 8(7), 878–885, Doi 10.5943/mycosphere/8/7/4 Abstract A new species, Antrodia monomitica sp. nov., is described and illustrated from China based on morphological characters and molecular evidence. It is characterized by producing annual, fragile and nodulose basidiomata, a monomitic hyphal system with clamp connections on generative hyphae, hyaline, thin-walled and fusiform to mango-shaped basidiospores (6–7.5 × 2.3– 3 µm), and causing a typical brown rot. In phylogenetic analysis inferred from ITS and nLSU rDNA sequences, the new species forms a distinct lineage in the Antrodia s. l., and has a close relationship with A. oleracea. Key words – Fomitopsidaceae – phylogenetic analysis – taxonomy – wood-decaying fungi Introduction Antrodia P. Karst., typified with Polyporus serpens Fr. (=Antrodia albida (Fr.) Donk (Donk 1960, Ryvarden 1991), is characterized by a resupinate to effused-reflexed growth habit, white or pale colour of the context, a dimitic hyphal system with clamp connections on generative hyphae, hyaline, thin-walled, cylindrical to very narrow ellipsoid basidiospores which are negative in Melzer’s reagent and Cotton Blue, and causing a brown rot (Ryvarden & Melo 2014). Antrodia is a highly heterogeneous genus which is closely related to Fomitopsis P. -

A Phylogenetic Overview of the Antrodia Clade (Basidiomycota, Polyporales)
Mycologia, 105(6), 2013, pp. 1391–1411. DOI: 10.3852/13-051 # 2013 by The Mycological Society of America, Lawrence, KS 66044-8897 A phylogenetic overview of the antrodia clade (Basidiomycota, Polyporales) Beatriz Ortiz-Santana1 phylogenetic studies also have recognized the genera Daniel L. Lindner Amylocystis, Dacryobolus, Melanoporia, Pycnoporellus, US Forest Service, Northern Research Station, Center for Sarcoporia and Wolfiporia as part of the antrodia clade Forest Mycology Research, One Gifford Pinchot Drive, (SY Kim and Jung 2000, 2001; Binder and Hibbett Madison, Wisconsin 53726 2002; Hibbett and Binder 2002; SY Kim et al. 2003; Otto Miettinen Binder et al. 2005), while the genera Antrodia, Botanical Museum, University of Helsinki, PO Box 7, Daedalea, Fomitopsis, Laetiporus and Sparassis have 00014, Helsinki, Finland received attention in regard to species delimitation (SY Kim et al. 2001, 2003; KM Kim et al. 2005, 2007; Alfredo Justo Desjardin et al. 2004; Wang et al. 2004; Wu et al. 2004; David S. Hibbett Dai et al. 2006; Blanco-Dios et al. 2006; Chiu 2007; Clark University, Biology Department, 950 Main Street, Worcester, Massachusetts 01610 Lindner and Banik 2008; Yu et al. 2010; Banik et al. 2010, 2012; Garcia-Sandoval et al. 2011; Lindner et al. 2011; Rajchenberg et al. 2011; Zhou and Wei 2012; Abstract: Phylogenetic relationships among mem- Bernicchia et al. 2012; Spirin et al. 2012, 2013). These bers of the antrodia clade were investigated with studies also established that some of the genera are molecular data from two nuclear ribosomal DNA not monophyletic and several modifications have regions, LSU and ITS. A total of 123 species been proposed: the segregation of Antrodia s.l. -

Draft Genome Sequence of a Monokaryotic
Draft genome sequence of a monokaryotic model brown-rot fungus Postia (Rhodonia) placenta SB12 Jill Gaskell, Phil Kersten, Luis Larrondo, Paulo Canessa, Diego Martínez, David Hibbett, Monika Schmoll, Christian Kubicek, Angel Martinez, Jagjit Yadav, et al. To cite this version: Jill Gaskell, Phil Kersten, Luis Larrondo, Paulo Canessa, Diego Martínez, et al.. Draft genome sequence of a monokaryotic model brown-rot fungus Postia (Rhodonia) placenta SB12. Genomics Data, Elsevier, 2017, 14, pp.21-23. 10.1016/j.gdata.2017.08.003. hal-01802856 HAL Id: hal-01802856 https://hal.archives-ouvertes.fr/hal-01802856 Submitted on 8 Jun 2018 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License Genomics Data 14 (2017) 21–23 Contents lists available at ScienceDirect Genomics Data journal homepage: www.elsevier.com/locate/gdata Data in Brief Draft genome sequence of a monokaryotic model brown-rot fungus Postia MARK (Rhodonia) placenta SB12 Jill Gaskella, Phil Kerstena, Luis F. Larrondob, Paulo Canessab,c, Diego Martinezd,1, David Hibbette, Monika Schmollf, Christian P. Kubicekg, Angel T. Martinezh, Jagjit Yadavi, Emma Masterj, Jon Karl Magnusonk, Debbie Yaverl, Randy Berkal, Kathleen Lailm, Cindy Chenm, Kurt LaButtim, Matt Nolanm, Anna Lipzenm, Andrea Aertsm, Robert Rileym, Kerrie Barrym, ⁎ Bernard Henrissatn,o,p,q, Robert Blanchetter, Igor V. -

Evidence of Subterranean Termite Feeding Deterrent Produced by Brown Rot Fungus Fibroporia Radiculosa (Peck) Parmasto 1968 (Polyporales, Fomitopsidaceae)
insects Article Evidence of Subterranean Termite Feeding Deterrent Produced by Brown Rot Fungus Fibroporia radiculosa (Peck) Parmasto 1968 (Polyporales, Fomitopsidaceae) Nadia Nuraniya Kamaluddin 1, Akiko Nakagawa-Izumi 1,*, Shota Nishizawa 1, Ayuko Fukunaga 1, Shuichi Doi 1, Tsuyoshi Yoshimura 2 and Sakae Horisawa 3 1 Graduate School of Life and Environmental Sciences, University of Tsukuba, Tsukuba 305-0872, Japan; [email protected](N.N.K.); [email protected], (S.N.); [email protected] (A.F.); [email protected] (S.D.) 2 Research Institute for Sustainable Humanosphere, Kyoto University, Uji, Kyoto 611-0011, Japan; [email protected] 3 School of Environmental Science and Engineering, Kochi University of Technology, Kami, Kochi 782-8502, Japan; [email protected] * Correspondence: [email protected]; Tel.: +81-29-853-4578 Academic Editor: Brian T. Forschler Received: 11 April 2016; Accepted: 9 August 2016; Published: 18 August 2016 Abstract: We found that decayed wood stakes with no termite damage collected from a termite-infested field exhibited a deterrent effect against the termite Reticulitermes speratus, Kolbe, 1885. The effect was observed to be lost or reduced by drying. After identification, it was found that the decayed stakes were infected by brown rot fungus Fibroporia radiculosa (Peck) Parmasto, 1968. In a no-choice feeding test, wood blocks decayed by this fungus under laboratory condition deterred R. speratus feeding and n-hexane extract from the decayed stake and blocks induced termite mortality. These data provided an insight into the interaction between wood-rot fungi and wood-feeding termites. -

Wood Research Indoor Fungal Destroyers of Wooden Materials – Their Identification in Present Review
WOOD RESEARCH 63 (2): 2018 203-214 INDOOR FUNGAL DESTROYERS OF WOODEN MATERIALS – THEIR IDENTIFICATION IN PRESENT REVIEW Ján Gáper Technical University in Zvolen, Faculty of Ecology and Environmental Sciences Department of Biology and Ecology Zvolen, Slovak Republic et University of Ostrava Faculty of Sciences, Department of Biology and Ecology Ostrava, Czech Republic Svetlana Gáperová, Terézia Gašparcová, Simona Kvasnová Matej Bel University, Faculty of Natural Sciences, Department of Biology and Ecology Banská Bystrica, Slovak Republic Peter Pristaš Pavol Jozef Šafárik University, Faculty of Natural Sciences Institute of Biology and Ecology Košice, Slovak Republic Kateřina Náplavová University of Ostrava, Faculty of Sciences, Department of Biology and Ecology Ostrava, Czech Republic et University of Minho Ceb-Centre of Biological Engineering, Campus De Gualtar Braga, Portugal (Received January 2018) ABSTRACT The wood-destroying fungi traditionally were separated from one another primarily on a basis of their sporocarp and/or strain morphology. Their diversity and simple macro- and 203 WOOD RESEARCH micromorphology of fungal structures have been major obstacles for more rapid progress in this regard. However, over the past two decades, there has been substantial progress in our understanding of genetic variability within traditionally recognized morphospecies. In this study we have overviewed genetic variation and phylogeography of macrofungi, which are important destroyers of wooden materials indoor of buildings. Several morphologically defined species of these fungal destroyers (Coniophora puteana, C. olivacea, C. arida, Serpula himantioides) have been shown to actually encompass several genetically isolated lineages (cryptic species). The protective efficacy against cryptic species within traditionally recognized morphospecies through laboratory tests (EN 113) and field trials (EN 252) might be sufficient to better prognosis of decay development in wooden materials for hazard assessment and for proper conservation and management plans. -

Phylogenetic Relationships of Antrodia Species and Related Taxa Based on Analyses of Nuclear Large Subunit Ribosomal DNA Sequences
Botanical Studies (2010) 51: 53-60. MicrobioLOGY Phylogenetic relationships of Antrodia species and related taxa based on analyses of nuclear large subunit ribosomal DNA sequences Zhi-HeYU1,Sheng-HuaWU2,*,Dong-MeiWANG3,andCheng-TauCHEN2 1College of Life Science, Yangtze University, Jingzhou 434025, Hubei, P.R. China 2Department of Botany, National Museum of Natural Science, Taichung 404, Taiwan 3Guangdong Provincial Key Laboratory of Microbial Culture Collection and Application, Guangdong Institute of Microbiology, Guangzhou 510070, P.R. China (Received March 24, 2008; Accepted July 10, 2009) ABSTRACT. ThisstudyaimedtoevaluaterelationshipsofAntrodia species and related taxa, including the taxonomic status of some Antrodiaspeciesthathavebeentreatedasseparategenera(Amyloporia,Fibroporia, andTaiwanofungus).AcomprehensivephylogeneticstudyofHomobasidiomycetespresentedbyBinderetal. in 2005, was consulted for sampling the taxa used for this analysis. The genera of the “residual” polyporoid cladeandphlebioidcladeoftheHomobasidiomyceteswerechosenasoutgroups,andthegenerabelonging totheAntrodiacladeandcorepolyporoidcladewereselectedasingroup.Phylogeneticanalysesofthisstudy werebasedonsequencedataderivedfromthenuclearlargesubunitribosomalDNA(nuc-LSUrDNA).The analytical methods of maximum parsimony (MP), maximum likelihood (ML), and Bayesian inference (BI) wereused.Resultsfromthesedifferentanalysesweregenerallyconsistent.Twomaincladeslackinghigh support were detected in the ingroup. Clade A consisted of taxa of the Antrodiaclade,includingalltwelve -

Phylogenetic Position and Taxonomy of Kusaghiporia Usambarensis Gen
Mycology An International Journal on Fungal Biology ISSN: 2150-1203 (Print) 2150-1211 (Online) Journal homepage: http://www.tandfonline.com/loi/tmyc20 Phylogenetic position and taxonomy of Kusaghiporia usambarensis gen. et sp. nov. (Polyporales) Juma Mahmud Hussein, Donatha Damian Tibuhwa & Sanja Tibell To cite this article: Juma Mahmud Hussein, Donatha Damian Tibuhwa & Sanja Tibell (2018): Phylogenetic position and taxonomy of Kusaghiporia usambarensis gen. et sp. nov. (Polyporales), Mycology, DOI: 10.1080/21501203.2018.1461142 To link to this article: https://doi.org/10.1080/21501203.2018.1461142 © 2018 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group. Published online: 15 Apr 2018. Submit your article to this journal View related articles View Crossmark data Full Terms & Conditions of access and use can be found at http://www.tandfonline.com/action/journalInformation?journalCode=tmyc20 MYCOLOGY, 2018 https://doi.org/10.1080/21501203.2018.1461142 Phylogenetic position and taxonomy of Kusaghiporia usambarensis gen. et sp. nov. (Polyporales) Juma Mahmud Husseina,b, Donatha Damian Tibuhwab and Sanja Tibella aInstitute of Organismal Biology, Evolutionary Biology Centre, Uppsala University, Uppsala, Sweden; bDepartment of Molecular Biology and Biotechnology, College of Natural & Applied Sciences, University of Dar es Salaam, Dar es Salaam, Tanzania ABSTRACT ARTICLE HISTORY A large polyporoid mushroom from the West Usambara Mountains in North-eastern Tanzania Received 17 November 2017 produces dark brown, up to 60-cm large fruiting bodies that at maturity may weigh more than Accepted 2 April 2018 10 kg. It has a high rate of mycelial growth and regeneration and was found growing on both dry KEYWORDS and green leaves of shrubs; attached to the base of living trees, and it was also observed to Kusaghiporia; molecular degrade dead snakes and insects accidentally coming into contact with it. -

CBD Fifth National Report
CONVENTION ON CONVENTION ON BIOLOGICAL DIVERSITY BIOLOGICAL DIVERSITY THE 5TH NATIONAL REPORT OF MONGOLIA biolohJA JJa folea YeehcO beiide& oa KnWWn}A. T HE CONVENTION ON BIOLOGI 5 T H N A T IO N AL R EPO RT C AL DIVERSITY OF M O N GOLIA MINISTRY OF ENVIRONMENT AND GREEN DEVELOPMENT STEPPE FORWARD PROGRAMME, Government building II, BIOLOGY DEPARTMENT, United Nation’s street 5/2, NATIONAL UNIVERSITY OF MONGOLIA TH Chingeltei District, Ulaanbaatar 15160, NUM, Building-2, Ulaanbaatar, Mongolia THE 5 NATIONAL REPORT OF Mongolia P.O.Box 537, Ulaanbaatar 210646A, Tel: 976-51-266197 Ulaanbaatar, Mongolia E-mail: [email protected] Tel: 976-99180148; 976-88305909; 976-88083058 MONGOLIA E-mail: [email protected]; [email protected]; [email protected] Designed by Mongolica Publishing 2014 Ulaanbaatar, Mongolia. 2014 CONVENTION ON BIOLOGICAL DIVERSITY CONVENTION ON BIOLOGICAL DIVERSITY FINANCED BY: MINISTRY OF ENVIRONMENT AND GREEN DEVELOPMENT CONVENTION ON BIOLOGICAL DIVERSITY-MONGOLIA GLOBAL ENVIRONMENT FACILITY UNITED NATIONS ENVIRONMENTAL PROGRAM CONVENTION ON BIOLOGICAL DIVERSITY THE 5TH NATIONAL REPORT OF MONGOLIA REPORT COMPILERS: COMPILED BY: S. GOMBOBAATAR STEPPE FORWARD PROGRAMME, NUM S. MYAGMARSUREN N. CONABOY М. Мunkhjargal TAXON COMPILERS: PLANT: B. OYUNTSETSEG, M. URGAMAL INVERTEBRATE: S. GANTIGMAA Fish, aMphibian, reptile: kh. Тerbish BIRD: S. GOMBOBAATAR MAMMAL: S. SHAR CONTRIBUTIONS FROM: EDITORS: NATIONAL UNIVERSITY OF MONGOLIA INSTITUTE OF BIOLOGY, MONGOLIAN ACADEMY OF SCIENCES D. BATBOLD MONGOLIAN ORNITHOLOGICAL SOCIETY -

Index to Volumes 1-20 (1950-1980)
Karstenia 20: 33- 79. 1981 Index to volumes 1-20 (1950-1980) Compiled by HILKKA KOPONEN, LIISA SEPPANEN and PENTTI ALANKO Editor's note nish Mycological Society, Sienilehti (earlier Sienitie toja-Svampnytt). Unfortunately the exact dates of issue could not be This first cumulative Index of Karstenia covers all the traced for the early numbers of Karstenia. For the papers so far published in the magazine. Karstenia, first eight volumes only the year of printing is known, dedicated to the famous Finnish mycologist Petter for volumes 9-14 we know the month and year of Adolf Karsten (1834-1917), was first published at ir printing. From vol. 15 onwards the exact date of pu regular intervals, between the years 1950-1976, and blication has been recorded. These dates are found on each issue was numbered as a separate volume (l-16). the covers of the issues, and are not repeated here. From volume 17 onwards, Karstenia has come out on It is hoped that this Index will prove a great help to an annual basis, each volume consisting of two issues those using the issues of Karstenia. As the Editor of (plus a supplement to vol. 18). Karstenia, I wish to express my best thanks to Mrs. During this time Karstenia has established its posi Hilkka Koponen, Lic.Phil., Miss Liisa Seppanen, tion as an important medium for the studies of Fin M.Sc., and Mr. Pentti Alanko, Head Gardener (all nish mycologists. Writers from abroad have contribu from the Department of Botany, University of Hel ted only occasionally, and mainly on topics in some sinki), who have painstakingly performed the labori way connected with Finland. -

Soppognyttevekster.No › Agarica-1998-Nr-24-25 T
-f 't),.. ~I:WI~TAD t'J'JfORHHMG l "International Mycological Directory" second edition 1990 av G.S.Hall & D.L.Hawkworth finner vi følgende om Fredrikstad Soppforening: MYCOWGICAL SOCIETY OF FREDRIKSTAD Status: Local Organisalion type: Amateur Society &ope: Specialist Conlact: Roy Kristiansen Addn!SS: Fredrikstad Soppforening, P.O. Box 167, N-1601 Fredrikstad, Norway. lnlen!sts: Edible fungi, macromycetes. Portrail: Frederikstad Soppforening was founded in 1973 and isopen to anyone interested in fungi. Its ai ms are to educate the public about edible and poisonous fungi and to improve knowledge of the regional non edible fungi. There are currently 130 subscribing members, represented by a biennially serving Board, consisting of a President, Vice-President, Treasurer, Secretary and three Members, who meet six to seven times per year. On average there are six membership meetings (usually two in the spring and four in the autumn) mainly devot ed to edible fungi, with lectures from Society members and occasionally from professionals. Five to six field trips are held in the season (including one in May), when an identification service for the general public is offered by authorized members who are trained in a University-based course. New species are deposited in the Herbaria at Oslo and Trondheim Universities. The Society offers to guide professionals and amateurs from other pans of Norway, and from other countries, through the region in search of special biotypes or races. MHtings: Occasional symposia are arranged on specific topics (eg Coninarius and Russula) by Society and outside specialists which attract panicipation from other Scandinavian countries. Publication: Journal: Agarica (ca 200 pages, two issues per year) is mainly dedicated to macrornycetes and accepts anicles written in Nordic languages, English, French or German.