Phylogenetic Position and Taxonomy of Kusaghiporia Usambarensis Gen
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
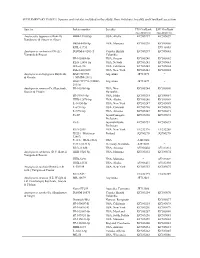
Suppl Table I
SUPPLEMENTARY TABLE I. Species and isolates included in the study, their vouchers, locality and GenBank accession Species Isolate number Locality ITS GenBank LSU GenBank accession no. accession no. Amylocystis lapponica (Romell) HHB-13400-Sp USA, Alaska KC585237 KC585059 Bondartsev & Singer ex Singer OKM-4418-Sp USA, Montana KC585238 KC585060 KHL-11755 − EU118603 Amyloporia carbonica (Overh.) DAOM-F-8281-T Canada, British KC585239 KC585061 Vampola & Pouzar Columbia FP-105585-Sp USA, Oregon KC585240 KC585062 RLG-12496-Sp USA, Nevada KC585241 KC585063 Wilcox-96 USA, California KC585242 KC585064 Zabel-40-GLN USA, New York KC585243 KC585065 Amyloporia nothofaginea Rajchenb. BAFC519794 Argentina JF713078 − & Gorjón (=MMBP-2011) BAFC519796 (=MBP- Argentina JF713079 − 2011a) Amyloporia sinuosa (Fr.) Rajchenb., FP-105386-Sp USA, New KC585244 KC585066 Gorjón & Pildain Hampshire FP-94464-Sp USA, Idaho KC585245 KC585067 HHB-12878-Sp USA, Alaska KC585246 KC585068 L-14130-Sp USA, New York KC585247 KC585069 L-6192-Sp USA, Colorado KC585248 KC585070 L-9792-Sp USA, Arizona KC585249 KC585071 Pa-3C JapanYamagata KC585250 KC585072 Prefecture Pa-3e JapanShizuoka KC585251 KC585073 Prefecture RLG-2538 USA, New York EU232196 EU232288 X725 (=Miettinen- Finland JQ700270 JQ700270 12407) P-115 (=RLG-2538) USA AJ416068 − P-211 (G-214) Germany, Karlsrube AJ345011 − RLG-1182R USA, Arizona AY966450 AY333831 Amyloporia sitchensis (D.V. Baxter) HHB-5320-Sp USA, Montana KC585252 KC585074 Vampola & Pouzar HHB-5298 USA, Montana − AY333829 HHB-12513 USA, Alaska AY966451 AY333830 -

Buglossoporus Pulvinus (Pers.) Donk) Входит В Число Редких Видов Макромицетов, Имеющих Высокий Природоохранный Статус Во Многих Странах Европы
УДК 582.284 : 581.527+581.42 (477.54) РЕДКИЙ ГРИБ PIPTOPORUS QUERCINUS (SCHRAD.) P. KARST. ИЗ НАЦИОНАЛЬНОГО ПРИРОДНОГО ПАРКА «ГОМОЛЬШАНСКИЕ ЛЕСА» Ордынец А.В., Акулов А.Ю. Кафедра микологии и фитоиммунологии, Харьковский национальный университет им. В.Н. Каразина пл. Свободы, 4, 61077, г. Харьков, Украина; e-mail: [email protected] Abstract. The article is devoted to a rare macromycete species Piptoporus quercinus (Schrad.) P. Karst., which has the high nature-conservative status and is included to Red lists of many European countries. The information about finding of this species on the territory of National nature park "Gomolshanskie lesa" (Zmiev area of Kharkiv district, Ukraine) is resulted. The localities of species detection on the territory of National park, the morphological, taxonomical and ecological characteristics of species, and the action plan for P. quercinus conservation are specified. Трутовый гриб Piptoporus quercinus (Schrad.) P. Karst. (=Buglossoporus pulvinus (Pers.) Donk) входит в число редких видов макромицетов, имеющих высокий природоохранный статус во многих странах Европы. Этот гриб развивается на старовозрастных дубах (Quercus spp.), и его численность существенно сокращается по мере уничтожения коренных дубрав [2; 5; 13; 15-19; 21; 22]. В Украине вид Piptoporus quercinus известен всего по нескольким находкам, и до недавнего времени был обнаружен на территории двух ботанико-географических регионов страны: Карпат и Закарпатья [1; 4]. В период 2002-2006 гг. этот вид был впервые обнаружен нами в Левобережной Украине: на валежных стволах и пнях Quercus robur L. в различных кварталах Национального природного парка «Гомольшанские леса». Учитывая редкость этого вида в общемировом масштабе, мы считаем необходимым привести подробную морфолого-таксономическую и экологическую характеристику этого вида, описать локалитеты его обнаружения на территории парка, а также мероприятия, необходимые для его сохранения. -

Diversity of Polyporales in the Malay Peninsular and the Application of Ganoderma Australe (Fr.) Pat
DIVERSITY OF POLYPORALES IN THE MALAY PENINSULAR AND THE APPLICATION OF GANODERMA AUSTRALE (FR.) PAT. IN BIOPULPING OF EMPTY FRUIT BUNCHES OF ELAEIS GUINEENSIS MOHAMAD HASNUL BIN BOLHASSAN FACULTY OF SCIENCE UNIVERSITY OF MALAYA KUALA LUMPUR 2013 DIVERSITY OF POLYPORALES IN THE MALAY PENINSULAR AND THE APPLICATION OF GANODERMA AUSTRALE (FR.) PAT. IN BIOPULPING OF EMPTY FRUIT BUNCHES OF ELAEIS GUINEENSIS MOHAMAD HASNUL BIN BOLHASSAN THESIS SUBMITTED IN FULFILMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY INSTITUTE OF BIOLOGICAL SCIENCES FACULTY OF SCIENCE UNIVERSITY OF MALAYA KUALA LUMPUR 2013 UNIVERSITI MALAYA ORIGINAL LITERARY WORK DECLARATION Name of Candidate: MOHAMAD HASNUL BIN BOLHASSAN (I.C No: 830416-13-5439) Registration/Matric No: SHC080030 Name of Degree: DOCTOR OF PHILOSOPHY Title of Project Paper/Research Report/Disertation/Thesis (“this Work”): DIVERSITY OF POLYPORALES IN THE MALAY PENINSULAR AND THE APPLICATION OF GANODERMA AUSTRALE (FR.) PAT. IN BIOPULPING OF EMPTY FRUIT BUNCHES OF ELAEIS GUINEENSIS. Field of Study: MUSHROOM DIVERSITY AND BIOTECHNOLOGY I do solemnly and sincerely declare that: 1) I am the sole author/writer of this work; 2) This Work is original; 3) Any use of any work in which copyright exists was done by way of fair dealing and for permitted purposes and any excerpt or extract from, or reference to or reproduction of any copyright work has been disclosed expressly and sufficiently and the title of the Work and its authorship have been acknowledge in this Work; 4) I do not have any actual -

Molecular Phylogeny of Laetiporus and Other Brown Rot Polypore Genera in North America
Mycologia, 100(3), 2008, pp. 417–430. DOI: 10.3852/07-124R2 # 2008 by The Mycological Society of America, Lawrence, KS 66044-8897 Molecular phylogeny of Laetiporus and other brown rot polypore genera in North America Daniel L. Lindner1 Key words: evolution, Fungi, Macrohyporia, Mark T. Banik Polyporaceae, Poria, root rot, sulfur shelf, Wolfiporia U.S.D.A. Forest Service, Madison Field Office of the extensa Northern Research Station, Center for Forest Mycology Research, One Gifford Pinchot Drive, Madison, Wisconsin 53726 INTRODUCTION The genera Laetiporus Murrill, Leptoporus Que´l., Phaeolus (Pat.) Pat., Pycnoporellus Murrill and Wolfi- Abstract: Phylogenetic relationships were investigat- poria Ryvarden & Gilb. contain species that possess ed among North American species of Laetiporus, simple septate hyphae, cause brown rots and produce Leptoporus, Phaeolus, Pycnoporellus and Wolfiporia annual, polyporoid fruiting bodies with hyaline using ITS, nuclear large subunit and mitochondrial spores. These shared morphological and physiologi- small subunit rDNA sequences. Members of these cal characters have been considered important in genera have poroid hymenophores, simple septate traditional polypore taxonomy (e.g. Gilbertson and hyphae and cause brown rots in a variety of substrates. Ryvarden 1986, Gilbertson and Ryvarden 1987, Analyses indicate that Laetiporus and Wolfiporia are Ryvarden 1991). However recent molecular work not monophyletic. All North American Laetiporus indicates that Laetiporus, Phaeolus and Pycnoporellus species formed a well supported monophyletic group fall within the ‘‘Antrodia clade’’ of true polypores (the ‘‘core Laetiporus clade’’ or Laetiporus s.s.) with identified by Hibbett and Donoghue (2001) while the exception of L. persicinus, which showed little Leptoporus and Wolfiporia fall respectively within the affinity for any genus for which sequence data are ‘‘phlebioid’’ and ‘‘core polyporoid’’ clades of true available. -

(2014), Volume 2, Issue 11, 238-245
ISSN 2320-5407 International Journal of Advanced Research (2014), Volume 2, Issue 11, 238-245 Journal homepage: http://www.journalijar.com INTERNATIONAL JOURNAL OF ADVANCED RESEARCH RESEARCH ARTICLE Diversity of family Meruliaceae from Jammu Division (J&K), India Jyoti*, Avneet Pal Singh & Gurpaul Singh Dhingra Department of Botany, Punjabi University, Patiala, 147002 India Manuscript Info Abstract Manuscript History: An account of eight resupinate, non-poroid taxa (Crustoderma corneum, Gyrophanopsis polonensis, Hyphoderma argillaceum, H. hjortstamii, H. Received: 25 September 2014 Final Accepted: 19 October 2014 setigerum, H. setigerum var. bicystidium, Hypochnicium wakefieldiae, Published Online: November 2014 Radulodon indicus) of family Meruliaceae (Class- Agaricomycetes, Phylum- Basidiomycota) has been given. All these are new reports for the Key words: Jammu Division in the state of Jammu and Kashmir (J&K). Of these, Basidiomycota, Agaricomycetes, Hyphoderma hjortstamii is a new record for India, Hypochnicium Meruliaceae. wakefieldiae new for the North Western Himalaya, Crustoderma corneum, Gyrophanopsis polonensis and H. setigerum var. bicystidium new for J&K. *Corresponding Author Jyoti Sharma Copy Right, IJAR, 2014,. All rights reserved Introduction While conducting fungal forays in the different localities of Jammu division in the state of Jammu and Kashmir (India), twelve collections of resupinate, non-poroid Agaricomycetous fungi were made. On the basis of comparison of macroscopic and microscopic features in the published literature (Thind & Rattan 1970, Eriksson & Ryvarden 1975, Rattan 1977, Eriksson & Ryvarden 1976, Eriksson et al. 1981, Wu SH. 1990, Stalpers 1998, Nakasone 2001, Bernicchia & Gorjón 2010), these have been identified as Crustoderma corneum, Gyrophanopsis polonensis, Hyphoderma argillaceum, H. hjortstamii, H. setigerum, H. setigerum var. bicystidium, Hypochnicium wakefieldiae and Radulodon indicus. -

Phylogenetic Classification of Trametes
TAXON 60 (6) • December 2011: 1567–1583 Justo & Hibbett • Phylogenetic classification of Trametes SYSTEMATICS AND PHYLOGENY Phylogenetic classification of Trametes (Basidiomycota, Polyporales) based on a five-marker dataset Alfredo Justo & David S. Hibbett Clark University, Biology Department, 950 Main St., Worcester, Massachusetts 01610, U.S.A. Author for correspondence: Alfredo Justo, [email protected] Abstract: The phylogeny of Trametes and related genera was studied using molecular data from ribosomal markers (nLSU, ITS) and protein-coding genes (RPB1, RPB2, TEF1-alpha) and consequences for the taxonomy and nomenclature of this group were considered. Separate datasets with rDNA data only, single datasets for each of the protein-coding genes, and a combined five-marker dataset were analyzed. Molecular analyses recover a strongly supported trametoid clade that includes most of Trametes species (including the type T. suaveolens, the T. versicolor group, and mainly tropical species such as T. maxima and T. cubensis) together with species of Lenzites and Pycnoporus and Coriolopsis polyzona. Our data confirm the positions of Trametes cervina (= Trametopsis cervina) in the phlebioid clade and of Trametes trogii (= Coriolopsis trogii) outside the trametoid clade, closely related to Coriolopsis gallica. The genus Coriolopsis, as currently defined, is polyphyletic, with the type species as part of the trametoid clade and at least two additional lineages occurring in the core polyporoid clade. In view of these results the use of a single generic name (Trametes) for the trametoid clade is considered to be the best taxonomic and nomenclatural option as the morphological concept of Trametes would remain almost unchanged, few new nomenclatural combinations would be necessary, and the classification of additional species (i.e., not yet described and/or sampled for mo- lecular data) in Trametes based on morphological characters alone will still be possible. -

Short-Read Sequencing for Genomic Analysis of the Brown Rot Fungus Fibroporia Radiculosa
Short-Read Sequencing for Genomic Analysis of the Brown Rot Fungus Fibroporia radiculosa Juliet D. Tang,a Andy D. Perkins,b Tad S. Sonstegard,c Steven G. Schroeder,c Shane C. Burgess,d and Susan V. Diehla Forest Products, Mississippi State University, Mississippi State, Mississippi, USAa; Computer Science and Engineering, Mississippi State University, Mississippi State, Mississippi, USAb; USDA ARS Bovine Functional Genomics Laboratory, Beltsville, Maryland, USAc; and Institute for Genomics, Biocomputing, and Biotechnology, Mississippi State University, Mississippi State, Mississippi, USAd The feasibility of short-read sequencing for genomic analysis was demonstrated for Fibroporia radiculosa, a copper-tolerant fungus that causes brown rot decay of wood. The effect of read quality on genomic assembly was assessed by filtering Illumina GAIIx reads from a single run of a paired-end library (75-nucleotide read length and 300-bp fragment size) at three different stringency levels and then assembling each data set with Velvet. A simple approach was devised to determine which filter strin- gency was “best.” Venn diagrams identified the regions containing reads that were used in an assembly but were of a low-enough quality to be removed by a filter. By plotting base quality histograms of reads in this region, we judged whether a filter was too stringent or not stringent enough. Our best assembly had a genome size of 33.6 Mb, an N50 of 65.8 kb for a k-mer of 51, and a maximum contig length of 347 kb. Using GeneMark, 9,262 genes were predicted. TargetP and SignalP analyses showed that among the 1,213 genes with secreted products, 986 had motifs for signal peptides and 227 had motifs for signal anchors. -

Climacodon Pulcherrimus a Badly Known Tropical Species, Present in Europe
Cryptogamie,Mycologie, 2007, 28 (1): 3-11 © 2007 Adac. Tous droits réservés Climacodon pulcherrimus a badly known tropical species, present in Europe Gabriel MORENOa*, María Natividad BLANCOa , Ibai OLARIAGAb &Julia CHECAa a Dpto. Biología Vegetal, Fac. Biología. Univ. de Alcalá, E-28871,Alcalá de Henares, Madrid. e-mail: [email protected] b Dpto. Biología Vegetal y Ecología (Botánica). Fac. Ciencias y Tecnología. Campus de Leioa. Univ. del País Vasco. Apartado 644. E-48080 Bilbao. Abstract – Climacodon pulcherrimus,apolymorphous species with a large distribution and habitat, is described macro- and microscopically. Its uncertain taxonomic position has led to the description of many synonymous species and placement in different genera. The type species of Hydnum pulcherrimum Berk. &M.A. Curtis is examined for the first time and it is compared with other collections from Malay Peninsula, Pakistan, USA and Spain. The study of Spanish collections enlarges the distribution to the South of Europe. Basidiomycota / Meruliaceae / Climacodon / Donkia / Hydnum / systematics / chorology / taxonomy INTRODUCTION In the last four years, we have collected some basidiomata of a saprophyte fungus with a basidioma of medium size, normally dimidiate with trametoid appearance and hydnoid hymenophore. Microscopically it is characterized by the presence of double or multiple clamp connections, ellipsoid basidiospores and absence of cystidia. Finally, we could determine it, with some difficulties, as a member of the genus Climacodon P. Karst., belonging to the family Meruliaceae P. Karst., order Polyporales Gäum. (Kirk et al., 2001). This genus includes species with conspicuous cystidia and hyphae with single clamp connections, with the only exception of C. pulcherrimus (Berk. -

A New Species of Antrodia (Basidiomycota, Polypores) from China
Mycosphere 8(7): 878–885 (2017) www.mycosphere.org ISSN 2077 7019 Article Doi 10.5943/mycosphere/8/7/4 Copyright © Guizhou Academy of Agricultural Sciences A new species of Antrodia (Basidiomycota, Polypores) from China Chen YY, Wu F* Institute of Microbiology, Beijing Forestry University, Beijing 100083, China Chen YY, Wu F 2017 –A new species of Antrodia (Basidiomycota, Polypores) from China. Mycosphere 8(7), 878–885, Doi 10.5943/mycosphere/8/7/4 Abstract A new species, Antrodia monomitica sp. nov., is described and illustrated from China based on morphological characters and molecular evidence. It is characterized by producing annual, fragile and nodulose basidiomata, a monomitic hyphal system with clamp connections on generative hyphae, hyaline, thin-walled and fusiform to mango-shaped basidiospores (6–7.5 × 2.3– 3 µm), and causing a typical brown rot. In phylogenetic analysis inferred from ITS and nLSU rDNA sequences, the new species forms a distinct lineage in the Antrodia s. l., and has a close relationship with A. oleracea. Key words – Fomitopsidaceae – phylogenetic analysis – taxonomy – wood-decaying fungi Introduction Antrodia P. Karst., typified with Polyporus serpens Fr. (=Antrodia albida (Fr.) Donk (Donk 1960, Ryvarden 1991), is characterized by a resupinate to effused-reflexed growth habit, white or pale colour of the context, a dimitic hyphal system with clamp connections on generative hyphae, hyaline, thin-walled, cylindrical to very narrow ellipsoid basidiospores which are negative in Melzer’s reagent and Cotton Blue, and causing a brown rot (Ryvarden & Melo 2014). Antrodia is a highly heterogeneous genus which is closely related to Fomitopsis P. -

Three Species of Wood-Decaying Fungi in <I>Polyporales</I> New to China
MYCOTAXON ISSN (print) 0093-4666 (online) 2154-8889 Mycotaxon, Ltd. ©2017 January–March 2017—Volume 132, pp. 29–42 http://dx.doi.org/10.5248/132.29 Three species of wood-decaying fungi in Polyporales new to China Chang-lin Zhaoa, Shi-liang Liua, Guang-juan Ren, Xiao-hong Ji & Shuanghui He* Institute of Microbiology, Beijing Forestry University, No. 35 Qinghuadong Road, Haidian District, Beijing 100083, P.R. China * Correspondence to: [email protected] Abstract—Three wood-decaying fungi, Ceriporiopsis lagerheimii, Sebipora aquosa, and Tyromyces xuchilensis, are newly recorded in China. The identifications were based on morphological and molecular evidence. The phylogenetic tree inferred from ITS+nLSU sequences of 49 species of Polyporales nests C. lagerheimii within the phlebioid clade, S. aquosa within the gelatoporia clade, and T. xuchilensis within the residual polyporoid clade. The three species are described and illustrated based on Chinese material. Key words—Basidiomycota, polypore, taxonomy, white rot fungus Introduction Wood-decaying fungi play a key role in recycling nutrients of forest ecosystems by decomposing cellulose, hemicellulose, and lignin of the plant cell walls (Floudas et al. 2015). Polyporales, a large order in Basidiomycota, includes many important genera of wood-decaying fungi. Recent molecular studies employing multi-gene datasets have helped to provide a phylogenetic overview of Polyporales, in which thirty-four valid families are now recognized (Binder et al. 2013). The diversity of wood-decaying fungi is very high in China because of the large landscape ranging from boreal to tropical zones. More than 1200 species of wood-decaying fungi have been found in China (Dai 2011, 2012), and some a Chang-lin Zhao and Shi-liang Liu contributed equally to this work and share first-author status 30 .. -

A Phylogenetic Overview of the Antrodia Clade (Basidiomycota, Polyporales)
Mycologia, 105(6), 2013, pp. 1391–1411. DOI: 10.3852/13-051 # 2013 by The Mycological Society of America, Lawrence, KS 66044-8897 A phylogenetic overview of the antrodia clade (Basidiomycota, Polyporales) Beatriz Ortiz-Santana1 phylogenetic studies also have recognized the genera Daniel L. Lindner Amylocystis, Dacryobolus, Melanoporia, Pycnoporellus, US Forest Service, Northern Research Station, Center for Sarcoporia and Wolfiporia as part of the antrodia clade Forest Mycology Research, One Gifford Pinchot Drive, (SY Kim and Jung 2000, 2001; Binder and Hibbett Madison, Wisconsin 53726 2002; Hibbett and Binder 2002; SY Kim et al. 2003; Otto Miettinen Binder et al. 2005), while the genera Antrodia, Botanical Museum, University of Helsinki, PO Box 7, Daedalea, Fomitopsis, Laetiporus and Sparassis have 00014, Helsinki, Finland received attention in regard to species delimitation (SY Kim et al. 2001, 2003; KM Kim et al. 2005, 2007; Alfredo Justo Desjardin et al. 2004; Wang et al. 2004; Wu et al. 2004; David S. Hibbett Dai et al. 2006; Blanco-Dios et al. 2006; Chiu 2007; Clark University, Biology Department, 950 Main Street, Worcester, Massachusetts 01610 Lindner and Banik 2008; Yu et al. 2010; Banik et al. 2010, 2012; Garcia-Sandoval et al. 2011; Lindner et al. 2011; Rajchenberg et al. 2011; Zhou and Wei 2012; Abstract: Phylogenetic relationships among mem- Bernicchia et al. 2012; Spirin et al. 2012, 2013). These bers of the antrodia clade were investigated with studies also established that some of the genera are molecular data from two nuclear ribosomal DNA not monophyletic and several modifications have regions, LSU and ITS. A total of 123 species been proposed: the segregation of Antrodia s.l. -

A Re-Evaluation of Neotropical Junghuhnia S.Lat. (Polyporales, Basidiomycota) Based on Morphological and Multigene Analyses
Persoonia 41, 2018: 130–141 ISSN (Online) 1878-9080 www.ingentaconnect.com/content/nhn/pimj RESEARCH ARTICLE https://doi.org/10.3767/persoonia.2018.41.07 A re-evaluation of Neotropical Junghuhnia s.lat. (Polyporales, Basidiomycota) based on morphological and multigene analyses M.C. Westphalen1,*, M. Rajchenberg2, M. Tomšovský3, A.M. Gugliotta1 Key words Abstract Junghuhnia is a genus of polypores traditionally characterised by a dimitic hyphal system with clamped generative hyphae and presence of encrusted skeletocystidia. However, recent molecular studies revealed that Mycodiversity Junghuhnia is polyphyletic and most of the species cluster with Steccherinum, a morphologically similar genus phylogeny separated only by a hydnoid hymenophore. In the Neotropics, very little is known about the evolutionary relation- Steccherinaceae ships of Junghuhnia s.lat. taxa and very few species have been included in molecular studies. In order to test the taxonomy proper phylogenetic placement of Neotropical species of this group, morphological and molecular analyses were carried out. Specimens were collected in Brazil and used for DNA sequence analyses of the internal transcribed spacer and the large subunit of the nuclear ribosomal RNA gene, the translation elongation factor 1-α gene, and the second largest subunit of RNA polymerase II gene. Herbarium collections, including type specimens, were studied for morphological comparison and to confirm the identity of collections. The molecular data obtained revealed that the studied species are placed in three different genera. Specimens of Junghuhnia carneola represent two distinct species that group in a lineage within the phlebioid clade, separated from Junghuhnia and Steccherinum, which belong to the residual polyporoid clade.