A New Species of Antrodia (Basidiomycota, Polypores) from China
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Five Polypore Species New to India
CZECH MYCOLOGY 72(2): 151–161, JULY 24, 2020 (ONLINE VERSION, ISSN 1805-1421) Five polypore species new to India 1 2 1 RAMANDEEP KAUR ,GURPREET KAUR ,AVNEET PAL SINGH *, 1 GURPAUL SINGH DHINGRA 1 Department of Botany, Punjabi University, Patiala, IN-147002, Punjab, India 2 Department of Agriculture, Khalsa College, Amritsar, IN-143002, Punjab, India *corresponding author: [email protected] Kaur R., Kaur G., Singh A.P., Dhingra G.S. (2020): Five polypore species new to India. – Czech Mycol. 72(2): 151–161. In continuation of the exploration of the diversity of polyporoid fungi in north-west India, five polypores identified as Antrodia leucaena, A. pulvinascens, Fomitiporia apiahyna, Inocutis ludoviciana and Inonotus venezuelicus are presented as new to India. These species are reported based on material collected from localities in the Sirmaur District (Himachal Pradesh) and Patiala District (Punjab). Descriptions, photographs and line drawings of the new records from India are provided. Key words: Agaricomycetes, Polyporales, Hymenochaetales, white-rot fungi, north-west Himalaya, Punjab. Article history: received 24 April 2020, revised 21 June 2020, accepted 29 June 2020, published on- line 24 July 2020. DOI: https://doi.org/10.33585/cmy.72202 Kaur R., Kaur G., Singh A.P., Dhingra G.S. (2020): Pět druhů chorošů nových pro Indii. – Czech Mycol. 72(2): 151–161. V rámci pokračujícího výzkumu diverzity chorošotvarých hub v severozápadní Indii bylo objeve- no pět druhů, které jsou nové pro Indii: Antrodia leucaena, A. pulvinascens, Fomitiporia apiahyna, Inocutis ludoviciana a Inonotus venezuelicus. Jejich výskyt byl zaznamenán a materiál sebrán na lo- kalitách v okresu Sirmaur (stát Himáčalpradéš) a okresu Patiala (stat Paňdžáb). -
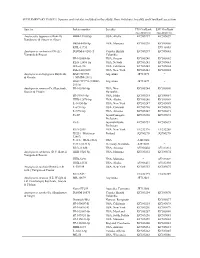
Suppl Table I
SUPPLEMENTARY TABLE I. Species and isolates included in the study, their vouchers, locality and GenBank accession Species Isolate number Locality ITS GenBank LSU GenBank accession no. accession no. Amylocystis lapponica (Romell) HHB-13400-Sp USA, Alaska KC585237 KC585059 Bondartsev & Singer ex Singer OKM-4418-Sp USA, Montana KC585238 KC585060 KHL-11755 − EU118603 Amyloporia carbonica (Overh.) DAOM-F-8281-T Canada, British KC585239 KC585061 Vampola & Pouzar Columbia FP-105585-Sp USA, Oregon KC585240 KC585062 RLG-12496-Sp USA, Nevada KC585241 KC585063 Wilcox-96 USA, California KC585242 KC585064 Zabel-40-GLN USA, New York KC585243 KC585065 Amyloporia nothofaginea Rajchenb. BAFC519794 Argentina JF713078 − & Gorjón (=MMBP-2011) BAFC519796 (=MBP- Argentina JF713079 − 2011a) Amyloporia sinuosa (Fr.) Rajchenb., FP-105386-Sp USA, New KC585244 KC585066 Gorjón & Pildain Hampshire FP-94464-Sp USA, Idaho KC585245 KC585067 HHB-12878-Sp USA, Alaska KC585246 KC585068 L-14130-Sp USA, New York KC585247 KC585069 L-6192-Sp USA, Colorado KC585248 KC585070 L-9792-Sp USA, Arizona KC585249 KC585071 Pa-3C JapanYamagata KC585250 KC585072 Prefecture Pa-3e JapanShizuoka KC585251 KC585073 Prefecture RLG-2538 USA, New York EU232196 EU232288 X725 (=Miettinen- Finland JQ700270 JQ700270 12407) P-115 (=RLG-2538) USA AJ416068 − P-211 (G-214) Germany, Karlsrube AJ345011 − RLG-1182R USA, Arizona AY966450 AY333831 Amyloporia sitchensis (D.V. Baxter) HHB-5320-Sp USA, Montana KC585252 KC585074 Vampola & Pouzar HHB-5298 USA, Montana − AY333829 HHB-12513 USA, Alaska AY966451 AY333830 -

Tree of Life Marula Oil in Africa
HerbalGram 79 • August – October 2008 HerbalGram 79 • August Herbs and Thyroid Disease • Rosehips for Osteoarthritis • Pelargonium for Bronchitis • Herbs of the Painted Desert The Journal of the American Botanical Council Number 79 | August – October 2008 Herbs and Thyroid Disease • Rosehips for Osteoarthritis • Pelargonium for Bronchitis • Herbs of the Painted Desert • Herbs of the Painted Bronchitis for Osteoarthritis Disease • Rosehips for • Pelargonium Thyroid Herbs and www.herbalgram.org www.herbalgram.org US/CAN $6.95 Tree of Life Marula Oil in Africa www.herbalgram.org Herb Pharm’s Botanical Education Garden PRESERVING THE FULL-SPECTRUM OF NATURE'S CHEMISTRY The Art & Science of Herbal Extraction At Herb Pharm we continue to revere and follow the centuries-old, time- proven wisdom of traditional herbal medicine, but we integrate that wisdom with the herbal sciences and technology of the 21st Century. We produce our herbal extracts in our new, FDA-audited, GMP- compliant herb processing facility which is located just two miles from our certified-organic herb farm. This assures prompt delivery of freshly-harvested herbs directly from the fields, or recently HPLC chromatograph showing dried herbs directly from the farm’s drying loft. Here we also biochemical consistency of 6 receive other organic and wildcrafted herbs from various parts of batches of St. John’s Wort extracts the USA and world. In producing our herbal extracts we use precision scientific instru- ments to analyze each herb’s many chemical compounds. However, You’ll find Herb Pharm we do not focus entirely on the herb’s so-called “active compound(s)” at fine natural products and, instead, treat each herb and its chemical compounds as an integrated whole. -

Phylogenetic Classification of Trametes
TAXON 60 (6) • December 2011: 1567–1583 Justo & Hibbett • Phylogenetic classification of Trametes SYSTEMATICS AND PHYLOGENY Phylogenetic classification of Trametes (Basidiomycota, Polyporales) based on a five-marker dataset Alfredo Justo & David S. Hibbett Clark University, Biology Department, 950 Main St., Worcester, Massachusetts 01610, U.S.A. Author for correspondence: Alfredo Justo, [email protected] Abstract: The phylogeny of Trametes and related genera was studied using molecular data from ribosomal markers (nLSU, ITS) and protein-coding genes (RPB1, RPB2, TEF1-alpha) and consequences for the taxonomy and nomenclature of this group were considered. Separate datasets with rDNA data only, single datasets for each of the protein-coding genes, and a combined five-marker dataset were analyzed. Molecular analyses recover a strongly supported trametoid clade that includes most of Trametes species (including the type T. suaveolens, the T. versicolor group, and mainly tropical species such as T. maxima and T. cubensis) together with species of Lenzites and Pycnoporus and Coriolopsis polyzona. Our data confirm the positions of Trametes cervina (= Trametopsis cervina) in the phlebioid clade and of Trametes trogii (= Coriolopsis trogii) outside the trametoid clade, closely related to Coriolopsis gallica. The genus Coriolopsis, as currently defined, is polyphyletic, with the type species as part of the trametoid clade and at least two additional lineages occurring in the core polyporoid clade. In view of these results the use of a single generic name (Trametes) for the trametoid clade is considered to be the best taxonomic and nomenclatural option as the morphological concept of Trametes would remain almost unchanged, few new nomenclatural combinations would be necessary, and the classification of additional species (i.e., not yet described and/or sampled for mo- lecular data) in Trametes based on morphological characters alone will still be possible. -

Short-Read Sequencing for Genomic Analysis of the Brown Rot Fungus Fibroporia Radiculosa
Short-Read Sequencing for Genomic Analysis of the Brown Rot Fungus Fibroporia radiculosa Juliet D. Tang,a Andy D. Perkins,b Tad S. Sonstegard,c Steven G. Schroeder,c Shane C. Burgess,d and Susan V. Diehla Forest Products, Mississippi State University, Mississippi State, Mississippi, USAa; Computer Science and Engineering, Mississippi State University, Mississippi State, Mississippi, USAb; USDA ARS Bovine Functional Genomics Laboratory, Beltsville, Maryland, USAc; and Institute for Genomics, Biocomputing, and Biotechnology, Mississippi State University, Mississippi State, Mississippi, USAd The feasibility of short-read sequencing for genomic analysis was demonstrated for Fibroporia radiculosa, a copper-tolerant fungus that causes brown rot decay of wood. The effect of read quality on genomic assembly was assessed by filtering Illumina GAIIx reads from a single run of a paired-end library (75-nucleotide read length and 300-bp fragment size) at three different stringency levels and then assembling each data set with Velvet. A simple approach was devised to determine which filter strin- gency was “best.” Venn diagrams identified the regions containing reads that were used in an assembly but were of a low-enough quality to be removed by a filter. By plotting base quality histograms of reads in this region, we judged whether a filter was too stringent or not stringent enough. Our best assembly had a genome size of 33.6 Mb, an N50 of 65.8 kb for a k-mer of 51, and a maximum contig length of 347 kb. Using GeneMark, 9,262 genes were predicted. TargetP and SignalP analyses showed that among the 1,213 genes with secreted products, 986 had motifs for signal peptides and 227 had motifs for signal anchors. -

A Phylogenetic Overview of the Antrodia Clade (Basidiomycota, Polyporales)
Mycologia, 105(6), 2013, pp. 1391–1411. DOI: 10.3852/13-051 # 2013 by The Mycological Society of America, Lawrence, KS 66044-8897 A phylogenetic overview of the antrodia clade (Basidiomycota, Polyporales) Beatriz Ortiz-Santana1 phylogenetic studies also have recognized the genera Daniel L. Lindner Amylocystis, Dacryobolus, Melanoporia, Pycnoporellus, US Forest Service, Northern Research Station, Center for Sarcoporia and Wolfiporia as part of the antrodia clade Forest Mycology Research, One Gifford Pinchot Drive, (SY Kim and Jung 2000, 2001; Binder and Hibbett Madison, Wisconsin 53726 2002; Hibbett and Binder 2002; SY Kim et al. 2003; Otto Miettinen Binder et al. 2005), while the genera Antrodia, Botanical Museum, University of Helsinki, PO Box 7, Daedalea, Fomitopsis, Laetiporus and Sparassis have 00014, Helsinki, Finland received attention in regard to species delimitation (SY Kim et al. 2001, 2003; KM Kim et al. 2005, 2007; Alfredo Justo Desjardin et al. 2004; Wang et al. 2004; Wu et al. 2004; David S. Hibbett Dai et al. 2006; Blanco-Dios et al. 2006; Chiu 2007; Clark University, Biology Department, 950 Main Street, Worcester, Massachusetts 01610 Lindner and Banik 2008; Yu et al. 2010; Banik et al. 2010, 2012; Garcia-Sandoval et al. 2011; Lindner et al. 2011; Rajchenberg et al. 2011; Zhou and Wei 2012; Abstract: Phylogenetic relationships among mem- Bernicchia et al. 2012; Spirin et al. 2012, 2013). These bers of the antrodia clade were investigated with studies also established that some of the genera are molecular data from two nuclear ribosomal DNA not monophyletic and several modifications have regions, LSU and ITS. A total of 123 species been proposed: the segregation of Antrodia s.l. -

Molecular Phylogeny and Taxonomic Position of Trametes Cervina and Description of a New Genus Trametopsis
CZECH MYCOL. 60(1): 1–11, 2008 Molecular phylogeny and taxonomic position of Trametes cervina and description of a new genus Trametopsis MICHAL TOMŠOVSKÝ Faculty of Forestry and Wood Technology, Mendel University of Agriculture and Forestry in Brno, Zemědělská 3, CZ-613 00, Brno, Czech Republic [email protected] Tomšovský M. (2008): Molecular phylogeny and taxonomic position of Trametes cervina and description of a new genus Trametopsis. – Czech Mycol. 60(1): 1–11. Trametes cervina (Schwein.) Bres. differs from other species of the genus by remarkable morpho- logical characters (shape of pores, hyphal system). Moreover, an earlier published comparison of the DNA sequences within the genus revealed considerable differences between this species and the re- maining European members of the genus Trametes. These results were now confirmed using se- quences of nuclear LSU and mitochondrial SSU regions of ribosomal DNA. The most related species of Trametes cervina are Ceriporiopsis aneirina and C. resinascens. According to these facts, the new genus Trametopsis Tomšovský is described and the new combination Trametopsis cervina (Schwein.) Tomšovský is proposed. Key words: Trametopsis, Trametes, ribosomal DNA, polypore, taxonomy. Tomšovský M. (2008): Molekulární fylogenetika a taxonomické zařazení outkovky jelení, Trametes cervina, a popis nového rodu Trametopsis. – Czech Mycol. 60(1): 1–11. Outkovka jelení, Trametes cervina (Schwein.) Bres., se liší od ostatních zástupců rodu nápadnými morfologickými znaky (tvar rourek, hyfový systém). Také dříve uveřejněné srovnání sekvencí DNA v rámci rodu Trametes odhalilo významné rozdíly mezi tímto druhem a ostatními evropskými zástupci rodu. Uvedené výsledky byly nyní potvrzeny za použití sekvencí jaderné LSU a mitochondriální SSU oblasti ribozomální DNA, přičemž nejpříbuznějšími druhu Trametes cervina jsou Ceriporiopsis anei- rina a C. -

A Preliminary Checklist of Arizona Macrofungi
A PRELIMINARY CHECKLIST OF ARIZONA MACROFUNGI Scott T. Bates School of Life Sciences Arizona State University PO Box 874601 Tempe, AZ 85287-4601 ABSTRACT A checklist of 1290 species of nonlichenized ascomycetaceous, basidiomycetaceous, and zygomycetaceous macrofungi is presented for the state of Arizona. The checklist was compiled from records of Arizona fungi in scientific publications or herbarium databases. Additional records were obtained from a physical search of herbarium specimens in the University of Arizona’s Robert L. Gilbertson Mycological Herbarium and of the author’s personal herbarium. This publication represents the first comprehensive checklist of macrofungi for Arizona. In all probability, the checklist is far from complete as new species await discovery and some of the species listed are in need of taxonomic revision. The data presented here serve as a baseline for future studies related to fungal biodiversity in Arizona and can contribute to state or national inventories of biota. INTRODUCTION Arizona is a state noted for the diversity of its biotic communities (Brown 1994). Boreal forests found at high altitudes, the ‘Sky Islands’ prevalent in the southern parts of the state, and ponderosa pine (Pinus ponderosa P.& C. Lawson) forests that are widespread in Arizona, all provide rich habitats that sustain numerous species of macrofungi. Even xeric biomes, such as desertscrub and semidesert- grasslands, support a unique mycota, which include rare species such as Itajahya galericulata A. Møller (Long & Stouffer 1943b, Fig. 2c). Although checklists for some groups of fungi present in the state have been published previously (e.g., Gilbertson & Budington 1970, Gilbertson et al. 1974, Gilbertson & Bigelow 1998, Fogel & States 2002), this checklist represents the first comprehensive listing of all macrofungi in the kingdom Eumycota (Fungi) that are known from Arizona. -

9B Taxonomy to Genus
Fungus and Lichen Genera in the NEMF Database Taxonomic hierarchy: phyllum > class (-etes) > order (-ales) > family (-ceae) > genus. Total number of genera in the database: 526 Anamorphic fungi (see p. 4), which are disseminated by propagules not formed from cells where meiosis has occurred, are presently not grouped by class, order, etc. Most propagules can be referred to as "conidia," but some are derived from unspecialized vegetative mycelium. A significant number are correlated with fungal states that produce spores derived from cells where meiosis has, or is assumed to have, occurred. These are, where known, members of the ascomycetes or basidiomycetes. However, in many cases, they are still undescribed, unrecognized or poorly known. (Explanation paraphrased from "Dictionary of the Fungi, 9th Edition.") Principal authority for this taxonomy is the Dictionary of the Fungi and its online database, www.indexfungorum.org. For lichens, see Lecanoromycetes on p. 3. Basidiomycota Aegerita Poria Macrolepiota Grandinia Poronidulus Melanophyllum Agaricomycetes Hyphoderma Postia Amanitaceae Cantharellales Meripilaceae Pycnoporellus Amanita Cantharellaceae Abortiporus Skeletocutis Bolbitiaceae Cantharellus Antrodia Trichaptum Agrocybe Craterellus Grifola Tyromyces Bolbitius Clavulinaceae Meripilus Sistotremataceae Conocybe Clavulina Physisporinus Trechispora Hebeloma Hydnaceae Meruliaceae Sparassidaceae Panaeolina Hydnum Climacodon Sparassis Clavariaceae Polyporales Gloeoporus Steccherinaceae Clavaria Albatrellaceae Hyphodermopsis Antrodiella -

Relationships Between Wood-Inhabiting Fungal Species
Silva Fennica 45(5) research articles SILVA FENNICA www.metla.fi/silvafennica · ISSN 0037-5330 The Finnish Society of Forest Science · The Finnish Forest Research Institute Relationships between Wood-Inhabiting Fungal Species Richness and Habitat Variables in Old-Growth Forest Stands in the Pallas-Yllästunturi National Park, Northern Boreal Finland Inari Ylläsjärvi, Håkan Berglund and Timo Kuuluvainen Ylläsjärvi, I., Berglund, H. & Kuuluvainen, T. 2011. Relationships between wood-inhabiting fungal species richness and habitat variables in old-growth forest stands in the Pallas-Yllästunturi National Park, northern boreal Finland. Silva Fennica 45(5): 995–1013. Indicators for biodiversity are needed for efficient prioritization of forests selected for conservation. We analyzed the relationships between 86 wood-inhabiting fungal (polypore) species richness and 35 habitat variables in 81 northern boreal old-growth forest stands in Finland. Species richness and the number of red-listed species were analyzed separately using generalized linear models. Most species were infrequent in the studied landscape and no species was encountered in all stands. The species richness increased with 1) the volume of coarse woody debris (CWD), 2) the mean DBH of CWD and 3) the basal area of living trees. The number of red-listed species increased along the same gradients, but the effect of basal area was not significant. Polypore species richness was significantly lower on western slopes than on flat topography. On average, species richness was higher on northern and eastern slopes than on western and southern slopes. The results suggest that a combination of habitat variables used as indicators may be useful in selecting forest stands to be set aside for polypore species conservation. -

Draft Genome Sequence of a Monokaryotic
Draft genome sequence of a monokaryotic model brown-rot fungus Postia (Rhodonia) placenta SB12 Jill Gaskell, Phil Kersten, Luis Larrondo, Paulo Canessa, Diego Martínez, David Hibbett, Monika Schmoll, Christian Kubicek, Angel Martinez, Jagjit Yadav, et al. To cite this version: Jill Gaskell, Phil Kersten, Luis Larrondo, Paulo Canessa, Diego Martínez, et al.. Draft genome sequence of a monokaryotic model brown-rot fungus Postia (Rhodonia) placenta SB12. Genomics Data, Elsevier, 2017, 14, pp.21-23. 10.1016/j.gdata.2017.08.003. hal-01802856 HAL Id: hal-01802856 https://hal.archives-ouvertes.fr/hal-01802856 Submitted on 8 Jun 2018 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License Genomics Data 14 (2017) 21–23 Contents lists available at ScienceDirect Genomics Data journal homepage: www.elsevier.com/locate/gdata Data in Brief Draft genome sequence of a monokaryotic model brown-rot fungus Postia MARK (Rhodonia) placenta SB12 Jill Gaskella, Phil Kerstena, Luis F. Larrondob, Paulo Canessab,c, Diego Martinezd,1, David Hibbette, Monika Schmollf, Christian P. Kubicekg, Angel T. Martinezh, Jagjit Yadavi, Emma Masterj, Jon Karl Magnusonk, Debbie Yaverl, Randy Berkal, Kathleen Lailm, Cindy Chenm, Kurt LaButtim, Matt Nolanm, Anna Lipzenm, Andrea Aertsm, Robert Rileym, Kerrie Barrym, ⁎ Bernard Henrissatn,o,p,q, Robert Blanchetter, Igor V. -

Ten Principles for Conservation Translocations of Threatened Wood- Inhabiting Fungi
Ten principles for conservation translocations of threatened wood- inhabiting fungi Jenni Nordén 1, Nerea Abrego 2, Lynne Boddy 3, Claus Bässler 4,5 , Anders Dahlberg 6, Panu Halme 7,8 , Maria Hällfors 9, Sundy Maurice 10 , Audrius Menkis 6, Otto Miettinen 11 , Raisa Mäkipää 12 , Otso Ovaskainen 9,13 , Reijo Penttilä 12 , Sonja Saine 9, Tord Snäll 14 , Kaisa Junninen 15,16 1Norwegian Institute for Nature Research, Gaustadalléen 21, NO-0349 Oslo, Norway. 2Dept of Agricultural Sciences, P.O. Box 27, FI-00014 University of Helsinki, Finland. 3Cardiff School of Biosciences, Sir Martin Evans Building, Museum Avenue, Cardiff CF10 3AX, UK 4Bavarian Forest National Park, D-94481 Grafenau, Germany. 5Technical University of Munich, Chair for Terrestrial Ecology, D-85354 Freising, Germany. 6Department of Forest Mycology and Plant Pathology, Swedish University of Agricultural Sciences, P.O.Box 7026, 750 07 Uppsala, Sweden. 7Department of Biological and Environmental Science, P.O. Box 35, FI-40014 University of Jyväskylä, Finland. 8School of Resource Wisdom, P.O. Box 35, FI-40014 University of Jyväskylä, Finland. 9Organismal and Evolutionary Biology Research Programme, P.O. Box 65, FI-00014 University of Helsinki, Finland. 10 Section for Genetics and Evolutionary Biology, University of Oslo, Blindernveien 31, 0316 Oslo, Norway. 11 Finnish Museum of Natural History, P.O. Box 7, FI-00014 University of Helsinki, Finland. 12 Natural Resources Institute Finland (Luke), Latokartanonkaari 9, FI-00790 Helsinki, Finland. 13 Centre for Biodiversity Dynamics, Department of Biology, Norwegian University of Science and Technology, N-7491 Trondheim, Norway. 14 Artdatabanken, Swedish University of Agricultural Sciences, P.O. Box 7007, SE-75007 Uppsala, Sweden.