Phylogenetic Relationships of Antrodia Species and Related Taxa Based on Analyses of Nuclear Large Subunit Ribosomal DNA Sequences
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
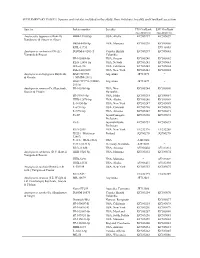
Suppl Table I
SUPPLEMENTARY TABLE I. Species and isolates included in the study, their vouchers, locality and GenBank accession Species Isolate number Locality ITS GenBank LSU GenBank accession no. accession no. Amylocystis lapponica (Romell) HHB-13400-Sp USA, Alaska KC585237 KC585059 Bondartsev & Singer ex Singer OKM-4418-Sp USA, Montana KC585238 KC585060 KHL-11755 − EU118603 Amyloporia carbonica (Overh.) DAOM-F-8281-T Canada, British KC585239 KC585061 Vampola & Pouzar Columbia FP-105585-Sp USA, Oregon KC585240 KC585062 RLG-12496-Sp USA, Nevada KC585241 KC585063 Wilcox-96 USA, California KC585242 KC585064 Zabel-40-GLN USA, New York KC585243 KC585065 Amyloporia nothofaginea Rajchenb. BAFC519794 Argentina JF713078 − & Gorjón (=MMBP-2011) BAFC519796 (=MBP- Argentina JF713079 − 2011a) Amyloporia sinuosa (Fr.) Rajchenb., FP-105386-Sp USA, New KC585244 KC585066 Gorjón & Pildain Hampshire FP-94464-Sp USA, Idaho KC585245 KC585067 HHB-12878-Sp USA, Alaska KC585246 KC585068 L-14130-Sp USA, New York KC585247 KC585069 L-6192-Sp USA, Colorado KC585248 KC585070 L-9792-Sp USA, Arizona KC585249 KC585071 Pa-3C JapanYamagata KC585250 KC585072 Prefecture Pa-3e JapanShizuoka KC585251 KC585073 Prefecture RLG-2538 USA, New York EU232196 EU232288 X725 (=Miettinen- Finland JQ700270 JQ700270 12407) P-115 (=RLG-2538) USA AJ416068 − P-211 (G-214) Germany, Karlsrube AJ345011 − RLG-1182R USA, Arizona AY966450 AY333831 Amyloporia sitchensis (D.V. Baxter) HHB-5320-Sp USA, Montana KC585252 KC585074 Vampola & Pouzar HHB-5298 USA, Montana − AY333829 HHB-12513 USA, Alaska AY966451 AY333830 -

Short-Read Sequencing for Genomic Analysis of the Brown Rot Fungus Fibroporia Radiculosa
Short-Read Sequencing for Genomic Analysis of the Brown Rot Fungus Fibroporia radiculosa Juliet D. Tang,a Andy D. Perkins,b Tad S. Sonstegard,c Steven G. Schroeder,c Shane C. Burgess,d and Susan V. Diehla Forest Products, Mississippi State University, Mississippi State, Mississippi, USAa; Computer Science and Engineering, Mississippi State University, Mississippi State, Mississippi, USAb; USDA ARS Bovine Functional Genomics Laboratory, Beltsville, Maryland, USAc; and Institute for Genomics, Biocomputing, and Biotechnology, Mississippi State University, Mississippi State, Mississippi, USAd The feasibility of short-read sequencing for genomic analysis was demonstrated for Fibroporia radiculosa, a copper-tolerant fungus that causes brown rot decay of wood. The effect of read quality on genomic assembly was assessed by filtering Illumina GAIIx reads from a single run of a paired-end library (75-nucleotide read length and 300-bp fragment size) at three different stringency levels and then assembling each data set with Velvet. A simple approach was devised to determine which filter strin- gency was “best.” Venn diagrams identified the regions containing reads that were used in an assembly but were of a low-enough quality to be removed by a filter. By plotting base quality histograms of reads in this region, we judged whether a filter was too stringent or not stringent enough. Our best assembly had a genome size of 33.6 Mb, an N50 of 65.8 kb for a k-mer of 51, and a maximum contig length of 347 kb. Using GeneMark, 9,262 genes were predicted. TargetP and SignalP analyses showed that among the 1,213 genes with secreted products, 986 had motifs for signal peptides and 227 had motifs for signal anchors. -

A New Species of Antrodia (Basidiomycota, Polypores) from China
Mycosphere 8(7): 878–885 (2017) www.mycosphere.org ISSN 2077 7019 Article Doi 10.5943/mycosphere/8/7/4 Copyright © Guizhou Academy of Agricultural Sciences A new species of Antrodia (Basidiomycota, Polypores) from China Chen YY, Wu F* Institute of Microbiology, Beijing Forestry University, Beijing 100083, China Chen YY, Wu F 2017 –A new species of Antrodia (Basidiomycota, Polypores) from China. Mycosphere 8(7), 878–885, Doi 10.5943/mycosphere/8/7/4 Abstract A new species, Antrodia monomitica sp. nov., is described and illustrated from China based on morphological characters and molecular evidence. It is characterized by producing annual, fragile and nodulose basidiomata, a monomitic hyphal system with clamp connections on generative hyphae, hyaline, thin-walled and fusiform to mango-shaped basidiospores (6–7.5 × 2.3– 3 µm), and causing a typical brown rot. In phylogenetic analysis inferred from ITS and nLSU rDNA sequences, the new species forms a distinct lineage in the Antrodia s. l., and has a close relationship with A. oleracea. Key words – Fomitopsidaceae – phylogenetic analysis – taxonomy – wood-decaying fungi Introduction Antrodia P. Karst., typified with Polyporus serpens Fr. (=Antrodia albida (Fr.) Donk (Donk 1960, Ryvarden 1991), is characterized by a resupinate to effused-reflexed growth habit, white or pale colour of the context, a dimitic hyphal system with clamp connections on generative hyphae, hyaline, thin-walled, cylindrical to very narrow ellipsoid basidiospores which are negative in Melzer’s reagent and Cotton Blue, and causing a brown rot (Ryvarden & Melo 2014). Antrodia is a highly heterogeneous genus which is closely related to Fomitopsis P. -

A Phylogenetic Overview of the Antrodia Clade (Basidiomycota, Polyporales)
Mycologia, 105(6), 2013, pp. 1391–1411. DOI: 10.3852/13-051 # 2013 by The Mycological Society of America, Lawrence, KS 66044-8897 A phylogenetic overview of the antrodia clade (Basidiomycota, Polyporales) Beatriz Ortiz-Santana1 phylogenetic studies also have recognized the genera Daniel L. Lindner Amylocystis, Dacryobolus, Melanoporia, Pycnoporellus, US Forest Service, Northern Research Station, Center for Sarcoporia and Wolfiporia as part of the antrodia clade Forest Mycology Research, One Gifford Pinchot Drive, (SY Kim and Jung 2000, 2001; Binder and Hibbett Madison, Wisconsin 53726 2002; Hibbett and Binder 2002; SY Kim et al. 2003; Otto Miettinen Binder et al. 2005), while the genera Antrodia, Botanical Museum, University of Helsinki, PO Box 7, Daedalea, Fomitopsis, Laetiporus and Sparassis have 00014, Helsinki, Finland received attention in regard to species delimitation (SY Kim et al. 2001, 2003; KM Kim et al. 2005, 2007; Alfredo Justo Desjardin et al. 2004; Wang et al. 2004; Wu et al. 2004; David S. Hibbett Dai et al. 2006; Blanco-Dios et al. 2006; Chiu 2007; Clark University, Biology Department, 950 Main Street, Worcester, Massachusetts 01610 Lindner and Banik 2008; Yu et al. 2010; Banik et al. 2010, 2012; Garcia-Sandoval et al. 2011; Lindner et al. 2011; Rajchenberg et al. 2011; Zhou and Wei 2012; Abstract: Phylogenetic relationships among mem- Bernicchia et al. 2012; Spirin et al. 2012, 2013). These bers of the antrodia clade were investigated with studies also established that some of the genera are molecular data from two nuclear ribosomal DNA not monophyletic and several modifications have regions, LSU and ITS. A total of 123 species been proposed: the segregation of Antrodia s.l. -

Relationships Between Wood-Inhabiting Fungal Species
Silva Fennica 45(5) research articles SILVA FENNICA www.metla.fi/silvafennica · ISSN 0037-5330 The Finnish Society of Forest Science · The Finnish Forest Research Institute Relationships between Wood-Inhabiting Fungal Species Richness and Habitat Variables in Old-Growth Forest Stands in the Pallas-Yllästunturi National Park, Northern Boreal Finland Inari Ylläsjärvi, Håkan Berglund and Timo Kuuluvainen Ylläsjärvi, I., Berglund, H. & Kuuluvainen, T. 2011. Relationships between wood-inhabiting fungal species richness and habitat variables in old-growth forest stands in the Pallas-Yllästunturi National Park, northern boreal Finland. Silva Fennica 45(5): 995–1013. Indicators for biodiversity are needed for efficient prioritization of forests selected for conservation. We analyzed the relationships between 86 wood-inhabiting fungal (polypore) species richness and 35 habitat variables in 81 northern boreal old-growth forest stands in Finland. Species richness and the number of red-listed species were analyzed separately using generalized linear models. Most species were infrequent in the studied landscape and no species was encountered in all stands. The species richness increased with 1) the volume of coarse woody debris (CWD), 2) the mean DBH of CWD and 3) the basal area of living trees. The number of red-listed species increased along the same gradients, but the effect of basal area was not significant. Polypore species richness was significantly lower on western slopes than on flat topography. On average, species richness was higher on northern and eastern slopes than on western and southern slopes. The results suggest that a combination of habitat variables used as indicators may be useful in selecting forest stands to be set aside for polypore species conservation. -

Piptoporus Betulinus, Fomitopsis Betulina)
Birch Fungi – Razor Strop, Birch Bracket (Piptoporus betulinus, Fomitopsis betulina) Features - This distinctive fungus only grows on birches, looks like nothing else that grows on birches, and is very common. It is not an aggressive tree killer, but is instead primarily in the business of decomposing dead trees. Birch polypore is present throughout the range of the birches, which grow around the globe in the northern hemisphere. The white-to-brownish fruiting bodies are annual, emerging from the bark of birches in spring and summer, but they deteriorate slowly and are still visible through the winter, though by then they have blackened and are not so attractive. Global Uses – Otzi the Iceman, who lived 5300 years ago, carried two fragments of a fruiting body of Fomitopsis betulina (formerly Piptoporus betulinus). Some scientists believe that Ӧtzi might have used the fungus for medical purposes and, although the idea arouses some controversy, the long tradition of the use of F. betulina in folk medicine is a fact. Infusion from F. betulina fruiting bodies was popular, especially in Russia, Baltic countries, Hungary, Romania for its nutritional and calming properties. Fungal tea was used against various cancer types, as an immunoenhancing, anti-parasitic agent, and a remedy for gastrointestinal disorders. Antiseptic and anti-bleeding dressings made from fresh F. betulina fruiting body were applied to wounds and the powder obtained from dried ones was used as a painkiller. Was used as a razor strop to sharpen fine edged blades. Medicinal Potential – Chemical Constituents: A and C, 1,3-beta-D-glucopyranan, B ergosta-7,22-dien-3- ol, fungisterol, ergosterol, agaric acid, dehydrotumulosic acid, ungalinic acid, betulinic acid, and tumulosic acid. -

Draft Genome Sequence of a Monokaryotic
Draft genome sequence of a monokaryotic model brown-rot fungus Postia (Rhodonia) placenta SB12 Jill Gaskell, Phil Kersten, Luis Larrondo, Paulo Canessa, Diego Martínez, David Hibbett, Monika Schmoll, Christian Kubicek, Angel Martinez, Jagjit Yadav, et al. To cite this version: Jill Gaskell, Phil Kersten, Luis Larrondo, Paulo Canessa, Diego Martínez, et al.. Draft genome sequence of a monokaryotic model brown-rot fungus Postia (Rhodonia) placenta SB12. Genomics Data, Elsevier, 2017, 14, pp.21-23. 10.1016/j.gdata.2017.08.003. hal-01802856 HAL Id: hal-01802856 https://hal.archives-ouvertes.fr/hal-01802856 Submitted on 8 Jun 2018 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License Genomics Data 14 (2017) 21–23 Contents lists available at ScienceDirect Genomics Data journal homepage: www.elsevier.com/locate/gdata Data in Brief Draft genome sequence of a monokaryotic model brown-rot fungus Postia MARK (Rhodonia) placenta SB12 Jill Gaskella, Phil Kerstena, Luis F. Larrondob, Paulo Canessab,c, Diego Martinezd,1, David Hibbette, Monika Schmollf, Christian P. Kubicekg, Angel T. Martinezh, Jagjit Yadavi, Emma Masterj, Jon Karl Magnusonk, Debbie Yaverl, Randy Berkal, Kathleen Lailm, Cindy Chenm, Kurt LaButtim, Matt Nolanm, Anna Lipzenm, Andrea Aertsm, Robert Rileym, Kerrie Barrym, ⁎ Bernard Henrissatn,o,p,q, Robert Blanchetter, Igor V. -

Evidence of Subterranean Termite Feeding Deterrent Produced by Brown Rot Fungus Fibroporia Radiculosa (Peck) Parmasto 1968 (Polyporales, Fomitopsidaceae)
insects Article Evidence of Subterranean Termite Feeding Deterrent Produced by Brown Rot Fungus Fibroporia radiculosa (Peck) Parmasto 1968 (Polyporales, Fomitopsidaceae) Nadia Nuraniya Kamaluddin 1, Akiko Nakagawa-Izumi 1,*, Shota Nishizawa 1, Ayuko Fukunaga 1, Shuichi Doi 1, Tsuyoshi Yoshimura 2 and Sakae Horisawa 3 1 Graduate School of Life and Environmental Sciences, University of Tsukuba, Tsukuba 305-0872, Japan; [email protected](N.N.K.); [email protected], (S.N.); [email protected] (A.F.); [email protected] (S.D.) 2 Research Institute for Sustainable Humanosphere, Kyoto University, Uji, Kyoto 611-0011, Japan; [email protected] 3 School of Environmental Science and Engineering, Kochi University of Technology, Kami, Kochi 782-8502, Japan; [email protected] * Correspondence: [email protected]; Tel.: +81-29-853-4578 Academic Editor: Brian T. Forschler Received: 11 April 2016; Accepted: 9 August 2016; Published: 18 August 2016 Abstract: We found that decayed wood stakes with no termite damage collected from a termite-infested field exhibited a deterrent effect against the termite Reticulitermes speratus, Kolbe, 1885. The effect was observed to be lost or reduced by drying. After identification, it was found that the decayed stakes were infected by brown rot fungus Fibroporia radiculosa (Peck) Parmasto, 1968. In a no-choice feeding test, wood blocks decayed by this fungus under laboratory condition deterred R. speratus feeding and n-hexane extract from the decayed stake and blocks induced termite mortality. These data provided an insight into the interaction between wood-rot fungi and wood-feeding termites. -

Polypore Diversity in North America with an Annotated Checklist
Mycol Progress (2016) 15:771–790 DOI 10.1007/s11557-016-1207-7 ORIGINAL ARTICLE Polypore diversity in North America with an annotated checklist Li-Wei Zhou1 & Karen K. Nakasone2 & Harold H. Burdsall Jr.2 & James Ginns3 & Josef Vlasák4 & Otto Miettinen5 & Viacheslav Spirin5 & Tuomo Niemelä 5 & Hai-Sheng Yuan1 & Shuang-Hui He6 & Bao-Kai Cui6 & Jia-Hui Xing6 & Yu-Cheng Dai6 Received: 20 May 2016 /Accepted: 9 June 2016 /Published online: 30 June 2016 # German Mycological Society and Springer-Verlag Berlin Heidelberg 2016 Abstract Profound changes to the taxonomy and classifica- 11 orders, while six other species from three genera have tion of polypores have occurred since the advent of molecular uncertain taxonomic position at the order level. Three orders, phylogenetics in the 1990s. The last major monograph of viz. Polyporales, Hymenochaetales and Russulales, accom- North American polypores was published by Gilbertson and modate most of polypore species (93.7 %) and genera Ryvarden in 1986–1987. In the intervening 30 years, new (88.8 %). We hope that this updated checklist will inspire species, new combinations, and new records of polypores future studies in the polypore mycota of North America and were reported from North America. As a result, an updated contribute to the diversity and systematics of polypores checklist of North American polypores is needed to reflect the worldwide. polypore diversity in there. We recognize 492 species of polypores from 146 genera in North America. Of these, 232 Keywords Basidiomycota . Phylogeny . Taxonomy . species are unchanged from Gilbertson and Ryvarden’smono- Wood-decaying fungus graph, and 175 species required name or authority changes. -

3. Overview of Existing Fungus Monitoring Protocols
Version 2.9, 20.12.2004 Monitoring protocol for wood-inhabiting fungi in the Alberta Biodiversity Monitoring Program for the Alberta Biodiversity Monitoring Program Jogeir N. Stokland 1 Anna-Liisa Sippola 2 December 2004 1Norwegian Institute of Land Inventory, Norway 2University of Lapland, Arctic Centre, Finland 1 Disclaimer The views, statements, and conclusions expressed in this report are those of the authors and should not be construed as conclusions or opinions of the ABMP. Development of the ABMP has continued since this report was produced. Thus, the report may not accurately reflect current ideas. 2 1. INTRODUCTION ...................................................................................................................... 4 1.1 Brief introduction to biodiversity in dead wood................................................................... 4 1.2 Knowledge about fungus diversity in Alberta and Canada .................................................. 5 2. FUNGAL DIVERSITY ON DEAD WOOD IN BOREAL FORESTS...................................... 5 2.1. Different fungus groups - their strengths and weaknesses for long-term monitoring ......... 5 2.2 Dead wood attributes that are important to fungal diversity............................................... 12 2.3 Brief comparison of fungi on dead wood in Scandinavia and Canada............................... 14 2.4 Brief description of Alberta forest types and expected fungus diversity patterns .............. 15 2.5 Polypore species diversity at local and regional levels...................................................... -

Universidade Federal De Santa Catarina Para a Obtenção Do Grau De Mestre Em Biologia De Fungos, Algas E Plantas
0 Gesieli Kaipper Figueiró ASPECTOS TAXONÔMICOS E FILOGENÉTICOS DE Antrodia s.l. COM A INCLUSÃO DE ESPÉCIMES DA REGIÃO NEOTROPICAL Dissertação submetida ao Programa de Pós Graduação em Biologia de Fungos, Algas e Plantas da Universidade Federal de Santa Catarina para a obtenção do Grau de mestre em Biologia de Fungos, Algas e Plantas. Orientador: Prof. Dr. Elisandro Ricardo Drechsler dos Santos. Coorientador: Dr. Gerardo Lucio Robledo. Florianópolis 2015 1 2 3 AGRADECIMENTOS Aos meus orientadores Ricardo e Gerardo, pelo aprendizado, confiança e disponibilidade durante esses dois anos, não tenho palavras para agradecê-los; Aos professores do programa de Pós-Graduação em Biologia de Fungos, Algas e Plantas pelo conhecimento transmitido durante a realização das disciplinas; Agradeço aos curadores dos Herbários CORD, FLOR, IBOT e URM, pelos empréstimos de materiais; Agradeço à Prof.ª Maria Alice pelo conhecimento transmitido e por todas as conversas e experiências no micolab; Ao Mateus Arduvino Reck pela amizade, aprendizado e disponibilidade para me ajudar sempre; Ao Dr. Aristóteles Góes Neto, que por meio do projeto “FungiBrBol” forneceu subsídios indispensáveis para a execução das análises moleculares deste trabalho; Agradeço imensamente a Val, Diogo e Carlos que tiveram toda a paciência do mundo me ensinando as coisas mais básicas e lindas sobre o mundo dos Fungos; Também sou grata aos colegas do Micolab pela amizade e carinho de sempre, compartilhando momentos de alegrias e tristezas, almoços de domingos e todas as datas comemorativas -

Wood Research Indoor Fungal Destroyers of Wooden Materials – Their Identification in Present Review
WOOD RESEARCH 63 (2): 2018 203-214 INDOOR FUNGAL DESTROYERS OF WOODEN MATERIALS – THEIR IDENTIFICATION IN PRESENT REVIEW Ján Gáper Technical University in Zvolen, Faculty of Ecology and Environmental Sciences Department of Biology and Ecology Zvolen, Slovak Republic et University of Ostrava Faculty of Sciences, Department of Biology and Ecology Ostrava, Czech Republic Svetlana Gáperová, Terézia Gašparcová, Simona Kvasnová Matej Bel University, Faculty of Natural Sciences, Department of Biology and Ecology Banská Bystrica, Slovak Republic Peter Pristaš Pavol Jozef Šafárik University, Faculty of Natural Sciences Institute of Biology and Ecology Košice, Slovak Republic Kateřina Náplavová University of Ostrava, Faculty of Sciences, Department of Biology and Ecology Ostrava, Czech Republic et University of Minho Ceb-Centre of Biological Engineering, Campus De Gualtar Braga, Portugal (Received January 2018) ABSTRACT The wood-destroying fungi traditionally were separated from one another primarily on a basis of their sporocarp and/or strain morphology. Their diversity and simple macro- and 203 WOOD RESEARCH micromorphology of fungal structures have been major obstacles for more rapid progress in this regard. However, over the past two decades, there has been substantial progress in our understanding of genetic variability within traditionally recognized morphospecies. In this study we have overviewed genetic variation and phylogeography of macrofungi, which are important destroyers of wooden materials indoor of buildings. Several morphologically defined species of these fungal destroyers (Coniophora puteana, C. olivacea, C. arida, Serpula himantioides) have been shown to actually encompass several genetically isolated lineages (cryptic species). The protective efficacy against cryptic species within traditionally recognized morphospecies through laboratory tests (EN 113) and field trials (EN 252) might be sufficient to better prognosis of decay development in wooden materials for hazard assessment and for proper conservation and management plans.