Sillago Sihama from Persian Gulf
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Traditional Ecological Knowledge in Andavadoaka, Southwest Madagascar
BLUE VENTURES CONSERVATION REPORT Josephine M. Langley Vezo Knowledge: Traditional Ecological Knowledge in Andavadoaka, southwest Madagascar 52 Avenue Road, London N6 5DR [email protected] Tel: +44 (0) 208 341 9819 Fax: +44 (0) 208 341 4821 BLUE VENTURES CONSERVATION REPORT Andavadoaka, 2006 © Blue Ventures 2006 Copyright in this publication and in all text, data and images contained herein, except as otherwise indicated, rests with Blue Ventures. Recommended citation: Langley, J. (2006). Vezo Knowledge: Traditional Ecological Knowledge in Andavadoaka, southwest Madagascar. BLUE VENTURES CONSERVATION REPORT Summary Many fisheries and marine conservation management (ii) Knowledge of marine resources is passed orally from projects throughout the world have been dogged by generation to generation failure as a result of a lack of acceptance of (iii) Traditional laws, taboos and ceremonies are commonly management interventions by local communities. used in natural resource management Evaluation of these failed studies has produced (iv) Lifestyle, food security and housing are all dependent extensive guidelines, manuals and new fields of study on natural resources and the use of coastal and marine (Bunce and Pomeroy, 2004). Community engagement, resources form an essential part of this participatory research, and promoting the use of local (v) The arrival of the Catholic Mission has reduced the knowledge have repeatedly emerged as steps necessary proportion of villagers adhering to traditional ancestor to address the problem of managing the development of worship people and their economies while simultaneously (vi) There has been a change in recent years from a barter protecting the environment (Berkes et al, 2001; Bunce and subsistence economy to a market-driven cash- based economy and Pomeroy, 2004; Wibera et al, 2004; Scholz et al, (vii) Increased income for some members of the 2004). -

Marine Biodiversity of the South East NRM Region
Marine Environment and Ecology Benthic Ecology Subprogram Marine Biodiversity of the South East NRM Region SARDI Publication No. F2009/000681-1 SARDI Research Report series No. 416 Keith Rowling, Shirley Sorokin, Leonardo Mantilla and David Currie SARDI Aquatic Sciences PO BOX 120 Henley Beach SA 5022 December 2009 Prepared for the Department for Environment and Heritage 1 Information Systems and Database Support Program Marine Biodiversity of the South East NRM Region Keith Rowling, Shirley Sorokin, Leonardo Mantilla and David Currie December 2009 SARDI Publication No. F2009/000681-1 SARDI Research Report Series No. 416 Prepared for the Department for Environment and Heritage 2 This Publication may be cited as: Rowling, K.P., Sorokin, S.J., Mantilla, L. & Currie, D.R.. (2009) Marine Biodiversity of the South East NRM Region. South Australian Research and Development Institute (Aquatic Sciences), Adelaide. SARDI Publication No. F2009/000681-1. South Australian Research and Development Institute SARDI Aquatic Sciences 2 Hamra Avenue West Beach SA 5024 Telephone: (08) 8207 5400 Facsimile: (08) 8207 5406 http://www.sardi.sa.gov.au DISCLAIMER The authors warrant that they have taken all reasonable care in producing this report. The report has been through the SARDI internal review process, and has been formally approved for release by the Chief of Division. Although all reasonable efforts have been made to ensure quality, SARDI does not warrant that the information in this report is free from errors or omissions. SARDI does not accept any liability for the contents of this report or for any consequences arising from its use or any reliance placed upon it. -

Final Report
FINAL REPORT REHABILITATION AND CONSERVATION THE SEAGRASS MEADOWS AT CAM HAI DONG, CAM RANH BAY, KHANH HOA PROVINCE, CENTRAL VIETNAM. Pham Huu Tri Institute of Oceanography Nhatrang, Vietnam NHATRANG, JUNE 2008 Table of Contents 1. INTRODUCTION: 3 2. OBJECTIVES: 4 3. MATERIALS AND METHODS: 4 3.1 Research methods: 4 3.2 Project sites: 4 3. 3 Duration of the project: 6 3.4 Planting techniques: 6 3.4.1 Seagrasses: 6 3.4.2 Sea-horse and sea-cucumber: 7 3.4.3 Methods of study the leaf growth rate and leaf production: 7 3.4.4 Management and taking care of the rehabilitated area at Cam Hai Dong, Cam Ranh ba: 7 4. RESULTS: 7 4.1 The natural environmental elements in the rehabilitated area – Cam Hai Dong- Cam Ranh: 7 4.2 Species diversity of seagrasses in the rehabilitated area: 8 4.3 Transplantation the Enhalus acoroides for planting: 9 4.3.1 For the shoots method: 9 4.3.2 For the seedlings methods: 11 4. 4 Results of restoration the sea-horse at the rehabilitated area: 13 4.5 Results of restoration the sea-cucumber at the rehabilitated area: 14 5. CONCLUSION: 15 6. RECOMMENDATION: 15 LITERATURE CITED: 16 APPENDIX: 17 2 1. INTRODUCTION: Khanh Hoa is the coastal province in southern central Vietnam, situated between 11°45’02’’ to 13°02’ 10’’ North latitude and from 108045’10’’to 109045’06’’ East longitude, the length of the coast lines about 300km. The shallow coastal area of Khanh Hoa province is often covered by seagrass beds especially in Cam Ranh bay area where the seagrass beds have concentrated distribution with the superficies up to more than 1000 ha, there are 6 common species of seagrasses have been found there, among them Enhalus acoroides which has long leaves (1-2 m), dense density (40- 100 shoots/m2) and high cover (70-100%) is dominant. -

Proceedings of the American Lobster Transferable Trap Workshop
Special Report No. 75 of the Atlantic States Marine Fisheries Commission Proceedings of the American Lobster Transferable Trap Workshop October 2002 Proceedings of the American Lobster Transferable Trap Workshop October 2002 Edited by Heather M. Stirratt Atlantic States Marine Fisheries Commission Convened by: Atlantic States Marine Fisheries Commission August 26, 2002 Washington, DC Proceedings of the American Lobster Transferable Trap Workshop ii Preface This document was prepared in cooperation with the Atlantic States Marine Fisheries Commission’s American Lobster Management Board, Technical Committee, Plan Development Team, Stock Assessment Subcommittee and the Advisory Panel. A report of the Atlantic States Marine Fisheries Commission pursuant to U.S. Department of Commerce, National Oceanic and Atmospheric Administration Award No. NA17FG2205. Proceedings of the American Lobster Transferable Trap Workshop iii Acknowledgments This report is the result of a Workshop on transferable trap programs for American lobster which was held on August 26, 2002, in Washington, DC. The workshop was convened and organized by a Workshop Steering Committee composed of: David Spencer (Advisory Panel, Chair), George Doll (New York Commercial Lobsterman, Advisory Panel Member), John Sorlien (Rhode Island Commercial Lobsterman, Advisory Panel Member), Todd Jesse (Massachusetts Commercial Lobsterman, Advisory Panel Member), Ernie Beckwith (Connecticut Department of Environmental Protection), Mark Gibson (Rhode Island Department of Environmental Management), -

Intrinsic Vulnerability in the Global Fish Catch
The following appendix accompanies the article Intrinsic vulnerability in the global fish catch William W. L. Cheung1,*, Reg Watson1, Telmo Morato1,2, Tony J. Pitcher1, Daniel Pauly1 1Fisheries Centre, The University of British Columbia, Aquatic Ecosystems Research Laboratory (AERL), 2202 Main Mall, Vancouver, British Columbia V6T 1Z4, Canada 2Departamento de Oceanografia e Pescas, Universidade dos Açores, 9901-862 Horta, Portugal *Email: [email protected] Marine Ecology Progress Series 333:1–12 (2007) Appendix 1. Intrinsic vulnerability index of fish taxa represented in the global catch, based on the Sea Around Us database (www.seaaroundus.org) Taxonomic Intrinsic level Taxon Common name vulnerability Family Pristidae Sawfishes 88 Squatinidae Angel sharks 80 Anarhichadidae Wolffishes 78 Carcharhinidae Requiem sharks 77 Sphyrnidae Hammerhead, bonnethead, scoophead shark 77 Macrouridae Grenadiers or rattails 75 Rajidae Skates 72 Alepocephalidae Slickheads 71 Lophiidae Goosefishes 70 Torpedinidae Electric rays 68 Belonidae Needlefishes 67 Emmelichthyidae Rovers 66 Nototheniidae Cod icefishes 65 Ophidiidae Cusk-eels 65 Trachichthyidae Slimeheads 64 Channichthyidae Crocodile icefishes 63 Myliobatidae Eagle and manta rays 63 Squalidae Dogfish sharks 62 Congridae Conger and garden eels 60 Serranidae Sea basses: groupers and fairy basslets 60 Exocoetidae Flyingfishes 59 Malacanthidae Tilefishes 58 Scorpaenidae Scorpionfishes or rockfishes 58 Polynemidae Threadfins 56 Triakidae Houndsharks 56 Istiophoridae Billfishes 55 Petromyzontidae -
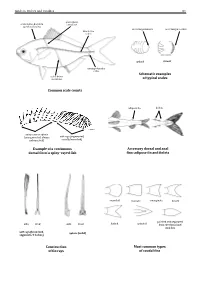
Field Identification Guide to the Living Marine Resources in Kenya
Guide to Orders and Families 81 lateral line scales above scales before dorsal fin outer margin smooth outer margin toothed (predorsal scales) lateral–line 114 scales cycloid ctenoidِّ scales circumpeduncular Schematic examples lateral line of typical scales scales below Common scale counts adipose fin finlets soft rays (segmented, spinyunbranched) rays or spines usually branched) (unsegmented, always Example of a continuous Accessory dorsal and anal dorsal fin of a spiny–rayed fish fins: adipose fin and finlets rounded truncate emarginate lunate side front side front from the dorsal and pointed and separated forked pointed soft rays (branched, spines (solid) segments, 2 halves) anal fins Construction Most common types of fin rays of caudal fins 82 Bony Fishes GUIDE TO ORDERS AND FAMILIES Order ELOPIFORMES – Tarpons and allies Fin spines absent; a single dorsal fin located above middle of body; pelvic fins in abdominal position; lateral line present; 23–25 branchiostegal rays; upper jaw extending past eye; tip of snout not overhanging mouth; colour silvery. ELOPIDAE Page 121 very small scales Ladyfishes To 90 cm. Coastal marine waters and estuaries; pelagic. A single species included in the Guide to Species.underside of head large mouth gular plate MEGALOPIDAE Page 121 last ray long Tarpons large scales To 55 cm. Coastal marine waters and estuaries; pelagic. A single species included in the Guide to Species.underside of head gular plate Order ALBULIFORMES – Bonefishes Fin spines absent; a single dorsal fin located above middle of body; pelvic fins in abdominal position; lateral line present; 6–16 branchiostegal rays; upper jaw not extending as far as front of eye; tip of snout overhanging mouth; colour silvery. -

5-Review-Fish-Habita
United Nations UNEP/GEF South China Sea Global Environment Environment Programme Project Facility UNEP/GEF/SCS/RWG-F.8/5 Date: 12th October 2006 Original: English Eighth Meeting of the Regional Working Group for the Fisheries Component of the UNEP/GEF Project: “Reversing Environmental Degradation Trends in the South China Sea and Gulf of Thailand” Bangka Belitung Province, Indonesia 1st - 4th November 2006 INFORMATION COLLATED BY THE FISHERIES AND HABITAT COMPONENTS OF THE SOUTH CHINA SEA PROJECT ON SITES IMPORTANT TO THE LIFE- CYCLES OF SIGNIFICANT FISH SPECIES UNEP/GEF/SCS/RWG-F.8/5 Page 1 IDENTIFICATION OF FISHERIES REFUGIA IN THE GULF OF THAILAND It was discussed at the Sixth Meeting of the Regional Scientific and Technical Committee (RSTC) in December 2006 that the Regional Working Group on Fisheries should take the following two-track approach to the identification of fisheries refugia: 1. Review known spawning areas for pelagic and invertebrate species, with the aim of evaluating these sites as candidate spawning refugia. 2. Evaluate each of the project’s habitat demonstration sites as potential juvenile/pre-recruit refugia for significant demersal species. Rationale for the Two-Track Approach to the Identification of Fisheries Refugia The two main life history events for fished species are reproduction and recruitment. It was noted by the RSTC that both of these events involve movement between areas, and some species, often pelagic fishes, migrate to particular spawning areas. It was also noted that many species also utilise specific coastal habitats such as coral reefs, seagrass, and mangroves as nursery areas. In terms of the effects of fishing, most populations of fished species are particularly vulnerable to the impacts of high levels of fishing effort in areas and at times where there are high abundances of (a) stock in spawning condition, (b) juveniles and pre-recruits, or (c) pre-recruits migrating to fishing grounds. -

Proceedings of the 40Th U.S.-Japan Aquaculture Panel Symposium
Hatchery Technology for High Quality Juvenile Production Proceedings of the 40th U.S.-Japan Aquaculture Panel Symposium University of Hawaii East West Center Honolulu, Hawaii October 22-23 2012 U.S. DEPARTMENT OF COMMERCE National Oceanic and Atmospheric Administration National Marine Fisheries Service NOAA Technical Memorandum NMFS-F/SPO-136 Hatchery Technology for High Quality Juvenile Production Proceedings of the 40th U.S.-Japan Aquaculture Panel Symposium University of Hawaii East West Center Honolulu, Hawaii October 22-23 2012 Mike Rust1, Paul Olin2, April Bagwill3, and Marie Fujitani3, editors 1Northwest Fisheries Science Center 2725 Montlake Boulevard East Seattle, Washington 98112 2California Sea Grant UCSD / Scripps Institution of Oceanography 133 Aviation Blvd., Suite 109 Santa Rosa CA 95403 3NOAA National Marine Fisheries Service 1315 East-West Highway Silver Spring, MD 20910 NOAA Technical Memorandum NMFS-F/SPO-136 December 2013 U.S. Department of Commerce Penny Pritzker, Secretary of Commerce National Oceanic and Atmospheric Administration Dr. Kathryn Sullivan, (Acting) NOAA Administrator National Marine Fisheries Service Samuel D. Rauch III, (Acting) Assistant Administrator for Fisheries SUGGESTED CITATION: Rust, M., P. Olin, A. Bagwill and M. Fujitani (editors). 2013. Hatchery Technology for High Quality Juvenile Production: Proceedings of the 40th U.S.-Japan Aquaculture Panel Symposium, Honolulu, Hawaii, October 22-23, 2012. U.S. Dept. Commerce, NOAA Tech. Memo. NMFS-F/SPO-136. A COPY OF THIS REPORT MAY BE OBTAINED FROM: Northwest Fisheries Science Center 2725 Montlake Boulevard East Seattle, Washington 98112 OR ONLINE AT: http://spo.nmfs.noaa.gov/tm/ Reference throughout this document to trade names does not imply endorsement by the National Marine Fisheries Service, NOAA. -

Global Progress in Ecosystem-Based Fisheries Management
26th Lowell Wakefield Fisheries Symposium Global Progress in Ecosystem-Based Fisheries Management editors gordon h. kruse • howard i. browman • kevern l. cochrane diana evans • glen s. jamieson • patricia a. livingston douglas woodby • chang ik zhang university of alaska fairbanks Global Progress in Ecosystem-Based Fisheries Management editors gordon h. kruse • howard i. browman • kevern l. cochrane diana evans • glen s. jamieson • patricia a. livingston douglas woodby • chang ik zhang Alaska university of alaska fairbanks Elmer E. Rasmuson Library Cataloging in Publication Data: Global progress in ecosystem-based fisheries management / editors : G.H. Kruse … [et al.] – Fairbanks, Alaska : Alaska Sea Grant College Program, University of Alaska Fairbanks, 2012. p. : ill. ; cm. – (Alaska Sea Grant College Program, University of Alaska Fairbanks ; AK-SG-12-01) Proceedings of the symposium Ecosystems 2010 : global progress on ecosystem- based fisheries management, November 8-11, 2010, Anchorage, Alaska. Includes bibliographical references. 1. Fishery management—Congresses. 2. Sustainable fisheries—Congresses. 3. Marine ecosystem management—Congresses. I. Title. II. Kruse, Gordon H. III. Series: Lowell Wakefield Fisheries symposia series (26th : 2010 : Anchorage, Alaska). IV. Series: Alaska Sea Grant College Program report ; AK-SG-12-01. SH329.S89 P76 2012 ISBN 978-1-56612-166-8 doi:10.4027/gpebfm.2012 Citation Kruse, G.H., H.I. Browman, K.L. Cochrane, D. Evans, G.S. Jamieson, P.A. Livingston, D. Woodby, and C.I. Zhang, eds. 2012. Global Progress in Ecosystem-Based Fisheries Management. Alaska Sea Grant, University of Alaska Fairbanks. Credits This book is published by Alaska Sea Grant, supported by the U.S. Department of Commerce, NOAA National Sea Grant Office, grant NA10OAR4170097, project A/161- 02, and by the University of Alaska Fairbanks with state funds. -

Field Identification Guide to the Living Marine Resources in Kenya
FAO FISH, FAO,FISH FINDER FINDER FAO SPECIES IDENTIFICATION GUIDE FOR FISHERY PURPOSES ISSN 1020-6868 FIELD IDENTIFICATION GUIDE TO THE LIVING FIELD IDENTIFICATION GUIDE TO THE LIVING GUIDE TO FIELD IDENTIFICATION MARINE RESOURCES OF KENYA MARINE RESOURCES OF KENYA MARINE RESOURCES This field guide covers the major resource groups likely to be encountered in the fisheries of Kenya. This includes shrimps, lobsters, crabs, bivalves, gastropods, cephalopods, sea cucumbers, sharks, batoid fishes, bony fishes, and sea turtles. Each resource group is introduced by a general section on technical terms and measurements pertinent to that group and an illustrated guide to orders and families of the group. The more important species are treated in a subsequent guide that includes scientific nomenclature, FAO names in English and French (were available), local names used in Kenya, diagnostic features, one or more illustrations, maximum size, and notes on fisheries and habitat. The guide is fully indexed and a list of further literature is appended. ' ISBN 978-92-5-107239-4 ISSN 1020-6868 e eTh\ II I I riC\ II I ! II A II" 9 789251 072394 I2741E/1/05.12 Cover: Illustration by Emanuela D’Antoni (FAO, Rome) FAO SPECIES IDENTIFICATION GUIDE FOR FISHERY PURPOSES FIELD IDENTIFICATION GUIDE TO THE LIVING MARINE RESOURCES OF KENYA by Rashid Anam Kenya Marine and Fisheries Research Institute (KMFRI) Mombasa, Kenya Edoardo Mostarda Fishery Resources Division FAO Fisheries Department Rome, Italy FOOD AND AGRICULTURE ORGANIZATION OF THE UNITED NATIONS Rome, 2012 The designations employed and the presentation of material in this information product do not imply the expression of any opinion whatsoever on the part of the Food and Agriculture Organization of the United Nations (FAO) concerning the legal or development status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. -

Biology, Stock Status and Management Summaries for Selected Fish Species in South-Western Australia
Fisheries Research Report No. 242, 2013 Biology, stock status and management summaries for selected fish species in south-western Australia Claire B. Smallwood, S. Alex Hesp and Lynnath E. Beckley Fisheries Research Division Western Australian Fisheries and Marine Research Laboratories PO Box 20 NORTH BEACH, Western Australia 6920 Correct citation: Smallwood, C. B.; Hesp, S. A.; and Beckley, L. E. 2013. Biology, stock status and management summaries for selected fish species in south-western Australia. Fisheries Research Report No. 242. Department of Fisheries, Western Australia. 180pp. Disclaimer The views and opinions expressed in this publication are those of the authors and do not necessarily reflect those of the Department of Fisheries Western Australia. While reasonable efforts have been made to ensure that the contents of this publication are factually correct, the Department of Fisheries Western Australia does not accept responsibility for the accuracy or completeness of the contents, and shall not be liable for any loss or damage that may be occasioned directly or indirectly through the use of, or reliance on, the contents of this publication. Fish illustrations Illustrations © R. Swainston / www.anima.net.au We dedicate this guide to the memory of our friend and colleague, Ben Chuwen Department of Fisheries 3rd floor SGIO Atrium 168 – 170 St Georges Terrace PERTH WA 6000 Telephone: (08) 9482 7333 Facsimile: (08) 9482 7389 Website: www.fish.wa.gov.au ABN: 55 689 794 771 Published by Department of Fisheries, Perth, Western Australia. Fisheries Research Report No. 242, March 2013. ISSN: 1035 - 4549 ISBN: 978-1-921845-56-7 ii Fisheries Research Report No.242, 2013 Contents ACKNOWLEDGEMENTS ............................................................................................... -
Recreational Fishing Guide 2021
Department of Primary Industries and Regional Development Recreational fishing guide 2021 New rules apply from 1 July 2021 see page 3 for details Includes Statewide bag and size limits for Western Australia, and Recreational Fishing from Boat Licence information Published June 2021 Page i Important disclaimer The Director General of the Department of Primary Industries and Regional Development (DPIRD) and the State of Western Australia accept no liability whatsoever by reason of negligence or otherwise arising from the use or release of this information or any part of it. This publication is to provide assistance or information. It is only a guide and does not replace the Fish Resources Management Act 1994 or the Fish Resources Management Regulations 1995. It cannot be used as a defence in a court of law. The information provided is current at the date of printing but may be subject to change. For the most up-to-date information on fishing and full details of legislation contact select DPIRD offices or visit dpird.wa.gov.au Copyright © State of Western Australia (Department of Primary Industries and Regional Development) 2021 Front cover photo: Tourism WA Department of Primary Industries and Regional Development Gordon Stephenson House, 140 William Street, Perth WA 6000 +61 1300 374 731 [email protected] dpird.wa.gov.au Page ii Contents Fish for the future .............................................2 Using this guide .................................................2 Changes to the rules – 2021 .............................3