Nuclear Domains of the RNA Subunit of Rnase P
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mouse Pop1 Conditional Knockout Project (CRISPR/Cas9)
https://www.alphaknockout.com Mouse Pop1 Conditional Knockout Project (CRISPR/Cas9) Objective: To create a Pop1 conditional knockout Mouse model (C57BL/6J) by CRISPR/Cas-mediated genome engineering. Strategy summary: The Pop1 gene (NCBI Reference Sequence: NM_152894 ; Ensembl: ENSMUSG00000022325 ) is located on Mouse chromosome 15. 17 exons are identified, with the ATG start codon in exon 2 and the TGA stop codon in exon 17 (Transcript: ENSMUST00000052290). Exon 4~5 will be selected as conditional knockout region (cKO region). Deletion of this region should result in the loss of function of the Mouse Pop1 gene. To engineer the targeting vector, homologous arms and cKO region will be generated by PCR using BAC clone RP23-365B13 as template. Cas9, gRNA and targeting vector will be co-injected into fertilized eggs for cKO Mouse production. The pups will be genotyped by PCR followed by sequencing analysis. Note: Exon 4 starts from about 9.92% of the coding region. The knockout of Exon 4~5 will result in frameshift of the gene. The size of intron 3 for 5'-loxP site insertion: 2480 bp, and the size of intron 5 for 3'-loxP site insertion: 1452 bp. The size of effective cKO region: ~2209 bp. The cKO region does not have any other known gene. Page 1 of 8 https://www.alphaknockout.com Overview of the Targeting Strategy Wildtype allele 5' gRNA region gRNA region 3' 1 4 5 6 7 17 Targeting vector Targeted allele Constitutive KO allele (After Cre recombination) Legends Exon of mouse Pop1 Homology arm cKO region loxP site Page 2 of 8 https://www.alphaknockout.com Overview of the Dot Plot Window size: 10 bp Forward Reverse Complement Sequence 12 Note: The sequence of homologous arms and cKO region is aligned with itself to determine if there are tandem repeats. -

Rpp2, an Essential Protein Subunit of Nuclear Rnase P, Is Required for Processing of Precursor Trnas and 35S Precursor Rrna in Saccharomyces Cerevisiae
Proc. Natl. Acad. Sci. USA Vol. 95, pp. 6716–6721, June 1998 Biochemistry Rpp2, an essential protein subunit of nuclear RNase P, is required for processing of precursor tRNAs and 35S precursor rRNA in Saccharomyces cerevisiae VIKTOR STOLC*, ALEXANDER KATZ†, AND SIDNEY ALTMAN†‡ †Department of Biology, Yale University, New Haven, CT 06520; and *Department of Cell Biology, Yale University School of Medicine, New Haven, CT 06510 Contributed by Sidney Altman, March 31, 1998 ABSTRACT RPP2, an essential gene that encodes a 15.8- has been suggested that RNase P is an ancestor of RNase kDa protein subunit of nuclear RNase P, has been identified MRP, because RNase MRP has been found only in eukaryotes in the genome of Saccharomyces cerevisiae. Rpp2 was detected (20). Yeast RNase MRP functions in processing of precursor by sequence similarity with a human protein, Rpp20, which rRNAs at the A3 site in the internal transcribed sequence of copurifies with human RNase P. Epitope-tagged Rpp2 can be the 35S precursor rRNA (21) and also cleaves RNA primers found in association with both RNase P and RNase mitochon- for mitochondrial DNA replication (22, 23). Although RNase drial RNA processing in immunoprecipitates from crude MRP does not cleave ptRNAs in vitro, its role in rRNA extracts of cells. Depletion of Rpp2 protein in vivo causes processing is affected by proteins that associate with RNase P accumulation of precursor tRNAs with unprocessed introns in vivo (11–14). RNase P functions in the biosynthesis of and 5* and 3* termini, and leads to defects in the processing tRNAs (8) and appears to have a role in rRNA processing in of the 35S precursor rRNA. -

Yeast Genome Gazetteer P35-65
gazetteer Metabolism 35 tRNA modification mitochondrial transport amino-acid metabolism other tRNA-transcription activities vesicular transport (Golgi network, etc.) nitrogen and sulphur metabolism mRNA synthesis peroxisomal transport nucleotide metabolism mRNA processing (splicing) vacuolar transport phosphate metabolism mRNA processing (5’-end, 3’-end processing extracellular transport carbohydrate metabolism and mRNA degradation) cellular import lipid, fatty-acid and sterol metabolism other mRNA-transcription activities other intracellular-transport activities biosynthesis of vitamins, cofactors and RNA transport prosthetic groups other transcription activities Cellular organization and biogenesis 54 ionic homeostasis organization and biogenesis of cell wall and Protein synthesis 48 plasma membrane Energy 40 ribosomal proteins organization and biogenesis of glycolysis translation (initiation,elongation and cytoskeleton gluconeogenesis termination) organization and biogenesis of endoplasmic pentose-phosphate pathway translational control reticulum and Golgi tricarboxylic-acid pathway tRNA synthetases organization and biogenesis of chromosome respiration other protein-synthesis activities structure fermentation mitochondrial organization and biogenesis metabolism of energy reserves (glycogen Protein destination 49 peroxisomal organization and biogenesis and trehalose) protein folding and stabilization endosomal organization and biogenesis other energy-generation activities protein targeting, sorting and translocation vacuolar and lysosomal -
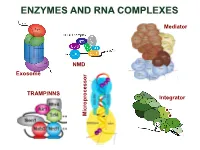
Enzymes and Rna Complexes
ENZYMES AND RNA COMPLEXES Mediator NMD Exosome NMD TRAMP/NNS Integrator Microprocessor RNA PROCESSING and DECAY machinery: RNases Protein Function Characteristics Exonucleases 5’ 3’ Xrn1 cytoplasmic, mRNA degradation processsive Rat1 nuclear, pre-rRNA, sn/snoRNA, pre-mRNA processing and degradation Rrp17/hNol12 nuclear, pre-rRNA processing Exosome 3’ 5’ multisubunit exo/endo complex subunits organized as in bacterial PNPase Rrp44/Dis3 catalytic subunit Exo/PIN domains, processsive Rrp4, Rrp40 pre-rRNA, sn/snoRNA processing, mRNA degradation Rrp41-43, 45-46 participates in NMD, ARE-dependent, non-stop decay Mtr3, Ski4 Mtr4 nuclear helicase cofactor DEAD box Rrp6 (Rrp47) nuclear exonuclease ( Rrp6 BP, cofactor) RNAse D homolog, processsive Ski2,3,7,8 cytoplasmic exosome cofactors. SKI complex helicase, GTPase Other 3’ 5’ Rex1-4 3’-5’ exonucleases, rRNA, snoRNA, tRNA processing RNase D homolog DXO 3’-5’ exonuclease in addition to decapping mtEXO 3’ 5’ mitochondrial degradosome RNA degradation in yeast Suv3/ Dss1 helicase/ 3’-5’ exonuclease DExH box/ RNase II homolog Deadenylation Ccr4/NOT/Pop2 major deadenylase complex (Ccr, Caf, Pop, Not proteins) Ccr4- Mg2+ dependent endonuclease Pan2p/Pan3 additional deadenylases (poliA tail length) RNase D homolog, poly(A) specific nuclease PARN mammalian deadenylase RNase D homolog, poly(A) specific nuclease Endonucleases RNase III -Rnt1 pre-rRNA, sn/snoRNA processing, mRNA degradation dsRNA specific -Dicer, Drosha siRNA/miRNA biogenesis, functions in RNAi PAZ, RNA BD, RNase III domains Ago2 Slicer -

A Disease-Linked Lncrna Mutation in Rnase MRP Inhibits Ribosome Synthesis
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.29.437572; this version posted March 29, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. A disease-linked lncRNA mutation in RNase MRP inhibits ribosome synthesis Nic Roberston1, Vadim Shchepachev1, David Wright2, Tomasz W. Turowski1, Christos Spanos1, Aleksandra Helwak1, Rose Zamoyska2, David Tollervey1 1 Wellcome Centre for Cell Biology, University of Edinburgh, Edinburgh, UK 2 Ashworth Laboratories, Institute of Immunology and Infection Research, University of Edinburgh, Edinburgh, UK Keywords: protein-RNA interaction; RNA-binding sites; UV crosslinking; mass spectrometry; genetic disease; Cartilage Hair Hypoplasia; ribosome synthesis; T cell activation Running title: RNase MRP defects cause ribosomopathy Highlights: • Mutations in RMRP lncRNA impair pre-rRNA processing and T cell activation • Patient derived fibroblasts show impaired pre-rRNA processing • Cells with the most common disease-linked mutation have specific processing defects • Cytoplasmic ribosomes and intact RNase MRP complexes are also reduced in these cells 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.03.29.437572; this version posted March 29, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Abstract Mutations in the human RMRP gene cause Cartilage Hair Hypoplasia (CHH), an autosomal recessive disorder characterized by skeletal abnormalities and impaired T cell activation. -

A Specialized Processing Body That Is Temporally and Asymmetrically Regulated During the Cell Cycle in Saccharomyces Cerevisiae
JCB: ARTICLE A specialized processing body that is temporally and asymmetrically regulated during the cell cycle in Saccharomyces cerevisiae Tina Gill, Jason Aulds, and Mark E. Schmitt Department of Biochemistry and Molecular Biology, State University of New York Upstate Medical University, Syracuse, NY 13210 Nase mitochondrial RNA processing (MRP) is an this spot was asymmetrically found in daughter cells, essential ribonucleoprotein endoribonuclease that where the RNase MRP substrate, CLB2 mRNA, localizes. R functions in the degradation of specifi c mRNAs in- Both the mitotic exit network and fourteen early ana- volved in cell cycle regulation. We have investigated phase release pathways are nonessential but important where this processing event occurs and how it is regu- for the temporal changes in localization. Asymmetric lo- lated. As expected, results demonstrate that RNase MRP calization was found to be dependent on the locasome. is predominantly localized in the nucleolus, where it The evidence suggests that these spots are specialized processes ribosomal RNAs. However, after the initiation processing bodies for the degradation of transcripts that of mitosis, RNase MRP localizes throughout the entire are cell cycle regulated and daughter cell localized. We nucleus and in a single discrete cytoplasmic spot that have called these TAM bodies for temporal asymmetric persists until the completion of telophase. Furthermore, MRP bodies. Introduction RNase mitochondrial RNA processing (MRP) is an essential ri- 27SA preribosomal RNA at the A3 site, forming the 5.8S(s) bonucleoprotein endoribonuclease that cleaves RNA substrates ribosomal RNA (rRNA; Schmitt and Clayton, 1993; Lygerou in a site-specifi c manner and is highly conserved in eukaryotes et al., 1996). -

Towards Reconstitution of Human Rnase P Protein Subunits with Human and Bacterial Rnase P Rnas
Towards reconstitution of human RNase P protein subunits with human and bacterial RNase P RNAs Research Thesis Presented in partial fulfillment of the requirements for graduation with research distinction in Biochemistry in the undergraduate colleges of The Ohio State University by Chigozirim Ekeke The Ohio State University June 2011 Project Advisor: Dr. Venkat Gopalan 1 DEDICATION Dedicated to the Ekeke family for their support, love, and blessings throughout my undergraduate career. 2 ACKNOWLEDGEMENTS I would like to thank Dr. Venkat Gopalan for his mentorship, wisdom, and unselfishness in helping me throughout my undergraduate research studies. He has taught me how to be a better scientist and student. I consider his presence in my life a true blessing. I thank Dr. Lien Lai for her guidance and willingness to help me throughout my research experience. Her opinions, critiques, and invaluable wisdom have molded me into a better researcher as well. I also thank Dr. Caroline Breitenberger for serving on my thesis committee and support throughout my collegiate career. I am grateful to the wonderful colleagues that I worked with in the Gopalan laboratory: Dr. Wen-Yi Chen, Dr. I-Ming Cho, Dr. Anil Challa, Dr. Gireesha Mohannath, Sathiyanarayanan Manivannan, and Cecilia Go. I appreciate their assistance and insights in helping me throughout my research. Additionally, I would like to thank Stella Lai, Emily Wong, Derek Smith, and Andrew Merriman for their sincere friendship and advice throughout this project. I am grateful and humbled by the financial support provided to me by the NSF Research Experience for Undergraduates Supplement (2010), OSU College of Biological Sciences (2009, 2010), and the OSU Colleges of the Arts and Sciences (2010-2011). -

Role and Regulation of the P53-Homolog P73 in the Transformation of Normal Human Fibroblasts
Role and regulation of the p53-homolog p73 in the transformation of normal human fibroblasts Dissertation zur Erlangung des naturwissenschaftlichen Doktorgrades der Bayerischen Julius-Maximilians-Universität Würzburg vorgelegt von Lars Hofmann aus Aschaffenburg Würzburg 2007 Eingereicht am Mitglieder der Promotionskommission: Vorsitzender: Prof. Dr. Dr. Martin J. Müller Gutachter: Prof. Dr. Michael P. Schön Gutachter : Prof. Dr. Georg Krohne Tag des Promotionskolloquiums: Doktorurkunde ausgehändigt am Erklärung Hiermit erkläre ich, dass ich die vorliegende Arbeit selbständig angefertigt und keine anderen als die angegebenen Hilfsmittel und Quellen verwendet habe. Diese Arbeit wurde weder in gleicher noch in ähnlicher Form in einem anderen Prüfungsverfahren vorgelegt. Ich habe früher, außer den mit dem Zulassungsgesuch urkundlichen Graden, keine weiteren akademischen Grade erworben und zu erwerben gesucht. Würzburg, Lars Hofmann Content SUMMARY ................................................................................................................ IV ZUSAMMENFASSUNG ............................................................................................. V 1. INTRODUCTION ................................................................................................. 1 1.1. Molecular basics of cancer .......................................................................................... 1 1.2. Early research on tumorigenesis ................................................................................. 3 1.3. Developing -

The Rnase MRP and Rnase P Complexes, Will Be Summarised
PDF hosted at the Radboud Repository of the Radboud University Nijmegen The following full text is a publisher's version. For additional information about this publication click this link. http://hdl.handle.net/2066/19147 Please be advised that this information was generated on 2021-09-28 and may be subject to change. The human The human RNase MRP complex RNase MRP complex Composition, assembly and role in human disease Hans van Eenennaam Hans van Eenennaam The human RNase MRP complex Composition, assembly and role in human disease Hans van Eenennaam, 2002 The human RNase MRP complex Composition, assembly and role in human disease een wetenschappelijke proeve op het gebied van de Natuurwetenschappen, Wiskunde en Informatica PROEFSCHRIFT ter verkrijging van de graad van doctor aan de Katholieke Universiteit Nijmegen, volgens besluit van het College van Decanen in het openbaar te verdedigen op vrijdag 14 juni 2002 des namiddags om 1:30 uur precies door Hans van Eenennaam Cover illustration Statue of the Dwarf Seneb and his family, painted limestone, height 34 cm, width 22.5 cm, geboren op 3 november 1973 Giza, Tomb of Seneb, Late Fifth - Early Sixth te Middelburg Dynasty. From: The Cairo museum Masterpieces of Egyptian Art, Francesco Tiradritti, Thames & Hudson Ltd, London, 1998 Promotor Prof. Dr. W.J. van Venrooij Co-promotor Dr. G.J.M. Pruijn voor mijn ouders Manuscriptcommissie Prof. Dr. L.B.A. van de Putte Prof. Dr. F.P.J.T. Rutjes Prof. Dr. H.F. Tabak (Universiteit van Amsterdam) ISBN 90-9015696-8 © 2002 by Hans van Eenennaam The research described in this thesis was performed at the Department of Biochemistry, Faculty of Science, University of Nijmegen, the Netherlands. -

A Novel Model for the Rnase MRP-Induced Switch Between the Different Forms of 5.8S Rrna
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 29 April 2021 doi:10.20944/preprints202104.0765.v1 Research article A novel model for the RNase MRP-induced switch between the different forms of 5.8S rRNA Xiao Li1,2,3, Janice M Zengel1 and Lasse Lindahl1 1 Department of Biological Sciences, University of Maryland Baltimore County (UMBC) 1000 Hilltop Circle, Baltimore, Maryland 21250, USA 2 Department of Biology, University of Rochester, Rochester, New York 14627, USA 3 Current address: Abbott Laboratories, San Diego, CA ribosome biogenesis; rRNA processing; RNase MRP; long/short 5.8S rRNA © 2021 by the author(s). Distributed under a Creative Commons CC BY license. Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 29 April 2021 doi:10.20944/preprints202104.0765.v1 Abstract: Processing of the RNA polymerase I pre-rRNA transcript into the mature 18S, 5.8S, and 25S rRNAs requires removing the “spacer” sequences. The canonical pathway for the removal of the ITS1 spacer, located between 18S and 5.8S rRNAs in the primary transcript, involves cleavages at the 3’ end of 18S rRNA and at two sites inside ITS1. The process generates a long and a short 5.8S rRNA that differ in the number of ITS1 nucleotides retained at the 5.8S 5’ end. Here we document a novel pathway that generates the long 5.8S for ITS1 while bypassing cleavage within ITS1. It entails a single endonuclease cut at the 3’-end of 18S rRNA followed by exonuclease Xrn1 degradation of ITS1. Mutations in RNase MRP increase the accumulation of long relative to short 5.8S rRNA; traditionally this is attributed to a decreased rate of RNase MRP cleavage at its target in ITS1, called A3. -

Subject Index Proc
13088 Subject Index Proc. Natl. Acad. Sci. USA 91 (1994) Apoptosis in substantia nigra following developmental striatal excitotoxic Brine shrimp injury, 8117 See Artemia Visualizing hippocampal synaptic function by optical detection of Ca2l Broccol entry through the N-methyl-D-aspartate channel, 8170 See Brassica Amygdala modulation of hippocampal-dependent and caudate Bromophenacyl bromide nucleus-dependent memory processes, 8477 Bromophenacyl bromide binding to the actin-bundling protein I-plastin Distribution of corticotropin-releasing factor receptor mRNA expression inhibits inositol trisphosphate-independent increase in Ca2l in human in the rat brain and pituitary, 8777 neutrophils, 3534 Brownian dynamics Preproenkephalin promoter yields region-specific and long-term Adhesion of hard spheres under the influence of double-layer, van der expression in adult brain after direct in vivo gene transfer via a Waals, and gravitational potentials at a solid/liquid interface, 3004 defective herpes simplex viral vector, 8979 Browsers Intravenous administration of a transferrin receptor antibody-nerve Thorn-like prickles and heterophylly in Cyanea: Adaptations to extinct growth factor conjugate prevents the degeneration of cholinergic avian browsers on Hawaii?, 2810 striatal neurons in a model of Huntington disease, 9077 Bruton agammaglobulinemia Axotomy induces the expression of vasopressin receptors in cranial and Genomic organization and structure of Bruton agammaglobulinemia spinal motor nuclei in the adult rat, 9636 tyrosine kinase: Localization -

Human Ribonuclease P: Subunits, Function, and Intranuclear Localization
Downloaded from rnajournal.cshlp.org on September 23, 2021 - Published by Cold Spring Harbor Laboratory Press RNA (2002), 8:1–7+ Cambridge University Press+ Printed in the USA+ Copyright © 2002 RNA Society+ DOI: 10+1017+S1355838201011189 REVIEW Human ribonuclease P: Subunits, function, and intranuclear localization NAYEF JARROUS Department of Molecular Biology, The Hebrew University–Hadassah Medical School, Jerusalem 91120, Israel ABSTRACT Catalytic complexes of nuclear ribonuclease P (RNase P) ribonucleoproteins are composed of several protein sub- units that appear to have specific roles in enzyme function in tRNA processing. This review describes recent progress made in the characterization of human RNase P, its relationship with the ribosomal RNA processing ribonucleoprotein RNase MRP, and the unexpected evolutionary conservation of its subunits. A new model for the biosynthesis of human RNase P is presented, in which this process is dynamic, transcription-dependent, and implicates functionally distinct nuclear compartments in tRNA biogenesis. Keywords: Cajal bodies; catalytic ribonucleoprotein; nucleolus; RNase MRP; RNase P; tRNA RIBONUCLEOPROTEIN COMPLEXES a ribonucleoprotein complex (Rossmanith et al+, 1995; OF RNase P Rossmanith & Karwan, 1998)+ These opposing find- ings provoke further debate on the biochemical nature Extensive purification analysis of RNase P from HeLa of mitochondrial and other organellar RNase P en- cells has revealed that the nuclear form of this tRNA zymes (Frank & Pace, 1998; Schon, 1999; Altman et al+,