Subject Index Proc
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Endogenous Reverse Transcriptase and Rnase H-Mediated Antiviral Mechanism in Embryonic Stem Cells
www.nature.com/cr www.cell-research.com ARTICLE Endogenous reverse transcriptase and RNase H-mediated antiviral mechanism in embryonic stem cells Junyu Wu1, Chunyan Wu1, Fan Xing1, Liu Cao1, Weijie Zeng1, Liping Guo1, Ping Li1, Yongheng Zhong1, Hualian Jiang1, Manhui Luo1, Guang Shi2, Lang Bu1, Yanxi Ji1, Panpan Hou1, Hong Peng1, Junjiu Huang2, Chunmei Li1 and Deyin Guo 1 Nucleic acid-based systems play important roles in antiviral defense, including CRISPR/Cas that adopts RNA-guided DNA cleavage to prevent DNA phage infection and RNA interference (RNAi) that employs RNA-guided RNA cleavage to defend against RNA virus infection. Here, we report a novel type of nucleic acid-based antiviral system that exists in mouse embryonic stem cells (mESCs), which suppresses RNA virus infection by DNA-mediated RNA cleavage. We found that the viral RNA of encephalomyocarditis virus can be reverse transcribed into complementary DNA (vcDNA) by the reverse transcriptase (RTase) encoded by endogenous retrovirus-like elements in mESCs. The vcDNA is negative-sense single-stranded and forms DNA/RNA hybrid with viral RNA. The viral RNA in the heteroduplex is subsequently destroyed by cellular RNase H1, leading to robust suppression of viral growth. Furthermore, either inhibition of the RTase activity or depletion of endogenous RNase H1 results in the promotion of virus proliferation. Altogether, our results provide intriguing insights into the antiviral mechanism of mESCs and the antiviral function of endogenized retroviruses and cellular RNase H. Such a natural nucleic acid-based antiviral mechanism in mESCs is referred to as ERASE (endogenous RTase/RNase H-mediated antiviral system), which is an addition to the previously known nucleic acid-based antiviral mechanisms including CRISPR/Cas in bacteria and RNAi in plants and invertebrates. -

Specific Principles of Genome-Wide RNA-Chromatin Interactions
ARTICLE There are amendments to this paper https://doi.org/10.1038/s41467-020-14337-6 OPEN RADICL-seq identifies general and cell type–specific principles of genome-wide RNA-chromatin interactions Alessandro Bonetti 1,2,18*, Federico Agostini 3,18, Ana Maria Suzuki1,4, Kosuke Hashimoto1, Giovanni Pascarella1, Juliette Gimenez 5, Leonie Roos6,7, Alex J. Nash6,7, Marco Ghilotti1, Christopher J. F. Cameron8,9, Matthew Valentine 1, Yulia A. Medvedeva10,11,12, Shuhei Noguchi1, Eneritz Agirre 2, Kaori Kashi1, Samudyata2, Joachim Luginbühl1, Riccardo Cazzoli13, Saumya Agrawal1, Nicholas M. Luscombe 3,14,15, Mathieu Blanchette8, Takeya Kasukawa 1, Michiel de Hoon1, Erik Arner1, 1234567890():,; Boris Lenhard 6,7,16, Charles Plessy 1, Gonçalo Castelo-Branco 2, Valerio Orlando5,17* & Piero Carninci 1* Mammalian genomes encode tens of thousands of noncoding RNAs. Most noncoding tran- scripts exhibit nuclear localization and several have been shown to play a role in the reg- ulation of gene expression and chromatin remodeling. To investigate the function of such RNAs, methods to massively map the genomic interacting sites of multiple transcripts have been developed; however, these methods have some limitations. Here, we introduce RNA And DNA Interacting Complexes Ligated and sequenced (RADICL-seq), a technology that maps genome-wide RNA–chromatin interactions in intact nuclei. RADICL-seq is a proximity ligation-based methodology that reduces the bias for nascent transcription, while increasing genomic coverage and unique mapping rate efficiency compared with existing methods. RADICL-seq identifies distinct patterns of genome occupancy for different classes of tran- scripts as well as cell type–specific RNA-chromatin interactions, and highlights the role of transcription in the establishment of chromatin structure. -

Activity of Selected Group of Monoterpenes in Alzheimer's
International Journal of Molecular Sciences Review Activity of Selected Group of Monoterpenes in Alzheimer’s Disease Symptoms in Experimental Model Studies—A Non-Systematic Review Karolina Wojtunik-Kulesza 1,*, Monika Rudkowska 2, Kamila Kasprzak-Drozd 1,* , Anna Oniszczuk 1 and Kinga Borowicz-Reutt 2 1 Department of Inorganic Chemistry, Medical University of Lublin, Chod´zki4a, 20-093 Lublin, Poland; [email protected] 2 Independent Experimental Neuropathophysiology Unit, Medical University of Lublin, Jaczewskiego 8b, 20-090 Lublin, Poland; [email protected] (M.R.); [email protected] (K.B.-R.) * Correspondence: [email protected] (K.W.-K.); [email protected] (K.K.-D.) Abstract: Alzheimer’s disease (AD) is the leading cause of dementia and cognitive function im- pairment. The multi-faced character of AD requires new drug solutions based on substances that incorporate a wide range of activities. Antioxidants, AChE/BChE inhibitors, BACE1, or anti-amyloid platelet aggregation substances are most desirable because they improve cognition with minimal side effects. Plant secondary metabolites, used in traditional medicine and pharmacy, are promising. Among these are the monoterpenes—low-molecular compounds with anti-inflammatory, antioxidant, Citation: Wojtunik-Kulesza, K.; enzyme inhibitory, analgesic, sedative, as well as other biological properties. The presented review Rudkowska, M.; Kasprzak-Drozd, K.; focuses on the pathophysiology of AD and a selected group of anti-neurodegenerative monoterpenes Oniszczuk, A.; Borowicz-Reutt, K. and monoterpenoids for which possible mechanisms of action have been explained. The main body Activity of Selected Group of of the article focuses on monoterpenes that have shown improved memory and learning, anxiolytic Monoterpenes in Alzheimer’s and sleep-regulating effects as determined by in vitro and in silico tests—followed by validation in Disease Symptoms in Experimental in vivo models. -

List of Union Reference Dates A
Active substance name (INN) EU DLP BfArM / BAH DLP yearly PSUR 6-month-PSUR yearly PSUR bis DLP (List of Union PSUR Submission Reference Dates and Frequency (List of Union Frequency of Reference Dates and submission of Periodic Frequency of submission of Safety Update Reports, Periodic Safety Update 30 Nov. 2012) Reports, 30 Nov. -

Yeast Genome Gazetteer P35-65
gazetteer Metabolism 35 tRNA modification mitochondrial transport amino-acid metabolism other tRNA-transcription activities vesicular transport (Golgi network, etc.) nitrogen and sulphur metabolism mRNA synthesis peroxisomal transport nucleotide metabolism mRNA processing (splicing) vacuolar transport phosphate metabolism mRNA processing (5’-end, 3’-end processing extracellular transport carbohydrate metabolism and mRNA degradation) cellular import lipid, fatty-acid and sterol metabolism other mRNA-transcription activities other intracellular-transport activities biosynthesis of vitamins, cofactors and RNA transport prosthetic groups other transcription activities Cellular organization and biogenesis 54 ionic homeostasis organization and biogenesis of cell wall and Protein synthesis 48 plasma membrane Energy 40 ribosomal proteins organization and biogenesis of glycolysis translation (initiation,elongation and cytoskeleton gluconeogenesis termination) organization and biogenesis of endoplasmic pentose-phosphate pathway translational control reticulum and Golgi tricarboxylic-acid pathway tRNA synthetases organization and biogenesis of chromosome respiration other protein-synthesis activities structure fermentation mitochondrial organization and biogenesis metabolism of energy reserves (glycogen Protein destination 49 peroxisomal organization and biogenesis and trehalose) protein folding and stabilization endosomal organization and biogenesis other energy-generation activities protein targeting, sorting and translocation vacuolar and lysosomal -

Download Supplementary
Supplementary Materials: High throughput virtual screening to discover inhibitors of the main protease of the coronavirus SARS-CoV-2 Olujide O. Olubiyi1,2*, Maryam Olagunju1, Monika Keutmann1, Jennifer Loschwitz1,3, and Birgit Strodel1,3* 1 Institute of Biological Information Processing: Structural Biochemistry, Forschungszentrum Jülich, Jülich, Germany 2 Department of Pharmaceutical Chemistry, Faculty of Pharmacy, Obafemi Awolowo University, Ile-Ife, Nigeria 3 Institute of Theoretical and Computational Chemistry, Heinrich Heine University Düsseldorf, 40225 Düsseldorf, Germany * Corresponding authors: [email protected], [email protected] List of Figures S1 Chemical fragments majorly featured in the top performing 9,515 synthetic com- pounds obtained from screening against the crystal structure of the SARS-CoV-2 main protease 3CLpro. .................................. 2 S2 Chemical fragments majorly featured in the top 2,102 synthetic compounds obtained from ensemble docking and application of cutoff values of ∆G ≤ −7.0 kcal/mol and ddyad ≤ 3.5 Å. ............................ 2 S3 The poses and 3CLpro–compound interactions of phthalocyanine and hypericin. 3 S4 The poses and 3CLpro–compound interactions of the four best non-FDA-approved and investigational drugs. ............................... 4 S5 The poses and 3CLpro–compound interactions of zeylanone and glabrolide. 5 List of Tables S1 Names and properties of the compounds binding best to the active site of 3CLpro. 6 1 Supporting Material: High throughput virtual screening for 3CLpro inhibitors Figure S1: Chemical fragments majorly featured in the top performing 9,515 synthetic com- pounds obtained from screening against the crystal structure of the SARS-CoV-2 main pro- tease 3CLpro. The numbers represent the occurrence in absolute numbers. -

Horizon Scanning Status Report June 2019
Statement of Funding and Purpose This report incorporates data collected during implementation of the Patient-Centered Outcomes Research Institute (PCORI) Health Care Horizon Scanning System, operated by ECRI Institute under contract to PCORI, Washington, DC (Contract No. MSA-HORIZSCAN-ECRI-ENG- 2018.7.12). The findings and conclusions in this document are those of the authors, who are responsible for its content. No statement in this report should be construed as an official position of PCORI. An intervention that potentially meets inclusion criteria might not appear in this report simply because the horizon scanning system has not yet detected it or it does not yet meet inclusion criteria outlined in the PCORI Health Care Horizon Scanning System: Horizon Scanning Protocol and Operations Manual. Inclusion or absence of interventions in the horizon scanning reports will change over time as new information is collected; therefore, inclusion or absence should not be construed as either an endorsement or rejection of specific interventions. A representative from PCORI served as a contracting officer’s technical representative and provided input during the implementation of the horizon scanning system. PCORI does not directly participate in horizon scanning or assessing leads or topics and did not provide opinions regarding potential impact of interventions. Financial Disclosure Statement None of the individuals compiling this information have any affiliations or financial involvement that conflicts with the material presented in this report. Public Domain Notice This document is in the public domain and may be used and reprinted without special permission. Citation of the source is appreciated. All statements, findings, and conclusions in this publication are solely those of the authors and do not necessarily represent the views of the Patient-Centered Outcomes Research Institute (PCORI) or its Board of Governors. -
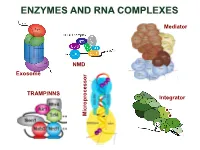
Enzymes and Rna Complexes
ENZYMES AND RNA COMPLEXES Mediator NMD Exosome NMD TRAMP/NNS Integrator Microprocessor RNA PROCESSING and DECAY machinery: RNases Protein Function Characteristics Exonucleases 5’ 3’ Xrn1 cytoplasmic, mRNA degradation processsive Rat1 nuclear, pre-rRNA, sn/snoRNA, pre-mRNA processing and degradation Rrp17/hNol12 nuclear, pre-rRNA processing Exosome 3’ 5’ multisubunit exo/endo complex subunits organized as in bacterial PNPase Rrp44/Dis3 catalytic subunit Exo/PIN domains, processsive Rrp4, Rrp40 pre-rRNA, sn/snoRNA processing, mRNA degradation Rrp41-43, 45-46 participates in NMD, ARE-dependent, non-stop decay Mtr3, Ski4 Mtr4 nuclear helicase cofactor DEAD box Rrp6 (Rrp47) nuclear exonuclease ( Rrp6 BP, cofactor) RNAse D homolog, processsive Ski2,3,7,8 cytoplasmic exosome cofactors. SKI complex helicase, GTPase Other 3’ 5’ Rex1-4 3’-5’ exonucleases, rRNA, snoRNA, tRNA processing RNase D homolog DXO 3’-5’ exonuclease in addition to decapping mtEXO 3’ 5’ mitochondrial degradosome RNA degradation in yeast Suv3/ Dss1 helicase/ 3’-5’ exonuclease DExH box/ RNase II homolog Deadenylation Ccr4/NOT/Pop2 major deadenylase complex (Ccr, Caf, Pop, Not proteins) Ccr4- Mg2+ dependent endonuclease Pan2p/Pan3 additional deadenylases (poliA tail length) RNase D homolog, poly(A) specific nuclease PARN mammalian deadenylase RNase D homolog, poly(A) specific nuclease Endonucleases RNase III -Rnt1 pre-rRNA, sn/snoRNA processing, mRNA degradation dsRNA specific -Dicer, Drosha siRNA/miRNA biogenesis, functions in RNAi PAZ, RNA BD, RNase III domains Ago2 Slicer -

(12) United States Patent (10) Patent No.: US 9,115,366 B2 Tissier Et Al
USOO9115366B2 (12) United States Patent (10) Patent No.: US 9,115,366 B2 Tissier et al. (45) Date of Patent: *Aug. 25, 2015 (54) SYSTEM FOR PRODUCING TERPENOIDS IN WO WO99,38957 * 8, 1999 .......... C12N 15/82 PLANTS WO WO99,38957 A1 8/1999 WO WOOOf 17327 A3 3.2000 Fre WO WO 01/20008 A2 3, 2001 (75) Inventors: Alain Tissier, Pertuis (FR); Christophe WO WO 2004/111183 A2 12/2004 Sallaud, Montpellier (FR); Denis WO WO 2006/04.0479 4/2006 Rontein,ontein, GreouxG les Bains (FR(FR) OTHER PUBLICATIONS (73) Assignee: PHILIP MORRIS PRODUCTS S.A., Aharoni, Aetal. The Plant Cell (Dec. 2003), vol. 15: pp. 2866-2884.* Neuchatel (CH) Besumbes, O. et al. Biotechnology and Bioengineering; Oct. 20. 2004; vol. 88, No. 2: pp. 168-175.* (*) Notice: Subject to any disclaimer, the term of this Wang, E. et al. Nature Biotechnology, Apr. 2001; vol. 19, pp. 371 patent is extended or adjusted under 35 37.4% U.S.C. 154(b) by 900 days. Wang, E. et al. Journal of Experimental Botany, Sep. 2002, vol. 53, No. 376; pp. 1891-1897.* This patent is Subject to a terminal dis Walker K. et al. Phytochemistry (2001) vol. 58; pp. 1-7.* claimer. Gutiérrez-Alcalá et al., A versatile promoter for the expression of proteins in glandular and non-glandular trichomes from a variety of plants, 56 J of Exp Botany No. 419, 2487-2494 (2005).* (21) Appl. No.: 11/814,943 Besumbes et al. (Metabolic Engineering of Isoprenoid Biosynthesis in Arabidopsis for the Production of Taxadiene, the First Committed (22) PCT Filed: Jan. -

A Disease-Linked Lncrna Mutation in Rnase MRP Inhibits Ribosome Synthesis
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.29.437572; this version posted March 29, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. A disease-linked lncRNA mutation in RNase MRP inhibits ribosome synthesis Nic Roberston1, Vadim Shchepachev1, David Wright2, Tomasz W. Turowski1, Christos Spanos1, Aleksandra Helwak1, Rose Zamoyska2, David Tollervey1 1 Wellcome Centre for Cell Biology, University of Edinburgh, Edinburgh, UK 2 Ashworth Laboratories, Institute of Immunology and Infection Research, University of Edinburgh, Edinburgh, UK Keywords: protein-RNA interaction; RNA-binding sites; UV crosslinking; mass spectrometry; genetic disease; Cartilage Hair Hypoplasia; ribosome synthesis; T cell activation Running title: RNase MRP defects cause ribosomopathy Highlights: • Mutations in RMRP lncRNA impair pre-rRNA processing and T cell activation • Patient derived fibroblasts show impaired pre-rRNA processing • Cells with the most common disease-linked mutation have specific processing defects • Cytoplasmic ribosomes and intact RNase MRP complexes are also reduced in these cells 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.03.29.437572; this version posted March 29, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Abstract Mutations in the human RMRP gene cause Cartilage Hair Hypoplasia (CHH), an autosomal recessive disorder characterized by skeletal abnormalities and impaired T cell activation. -

Pharmacokinetics, Pharmacodynamics and Metabolism Of
PHARMACOKINETICS, PHARMACODYNAMICS AND METABOLISM OF GTI-2040, A PHOSPHOROTHIOATE OLIGONUCLEOTIDE TARGETING R2 SUBUNIT OF RIBONUCLEOTIDE REDUCTASE DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Xiaohui Wei, M.S. * * * * * * The Ohio State University 2006 Approved by Dissertation Committee: Dr. Kenneth K. Chan, Adviser Adviser Dr. Guido Marcucci, Co-adviser Graduate Program in Pharmacy Dr. Thomas D. Schmittgen Dr. Robert J. Lee Co-Adviser Graduate Program in Pharmacy ABSTRACT Over the last several decades, antisense therapy has been developed into a promising gene-targeted strategy to specifically inhibit the gene expression. Ribonucleotide reductase (RNR), composing of subunits R1 and R2, is an important enzyme involved in the synthesis of all of the precursors used in DNA replication. Over- expression of R2 has been found in almost every type of cancer studied. GTI-2040 is a 20-mer phosphorothioate oligonucleotide targeting the coding region in mRNA of the R2 component of human RNR. In this project, clinical pharamcokinetics (PK), pharmacodynamics (PD) and metabolism of this novel therapeutics were investigated in patients with acute myeloid leukemia (AML). A picomolar specific hybridization-ligation ELISA method has been developed and validated for quantification of GTI-2040. GTI-2040 and neophectin complex was found to enhance drug cellular uptake and exhibited sequence- and dose-dependent down-regulation of R2 mRNA and protein in K562 cells. Robust intracellular concentrations (ICs) of GTI-2040 were achieved in peripheral blood mononuclear cells (PBMC) and bone marrow (BM) cells from treated AML patients. GTI-2040 concentrations in the nucleus of BM cells were found to correlate with the R2 mRNA down-regulation and disease response. -

Supplemental Methods
Supplemental Methods: Sample Collection Duplicate surface samples were collected from the Amazon River plume aboard the R/V Knorr in June 2010 (4 52.71’N, 51 21.59’W) during a period of high river discharge. The collection site (Station 10, 4° 52.71’N, 51° 21.59’W; S = 21.0; T = 29.6°C), located ~ 500 Km to the north of the Amazon River mouth, was characterized by the presence of coastal diatoms in the top 8 m of the water column. Sampling was conducted between 0700 and 0900 local time by gently impeller pumping (modified Rule 1800 submersible sump pump) surface water through 10 m of tygon tubing (3 cm) to the ship's deck where it then flowed through a 156 µm mesh into 20 L carboys. In the lab, cells were partitioned into two size fractions by sequential filtration (using a Masterflex peristaltic pump) of the pre-filtered seawater through a 2.0 µm pore-size, 142 mm diameter polycarbonate (PCTE) membrane filter (Sterlitech Corporation, Kent, CWA) and a 0.22 µm pore-size, 142 mm diameter Supor membrane filter (Pall, Port Washington, NY). Metagenomic and non-selective metatranscriptomic analyses were conducted on both pore-size filters; poly(A)-selected (eukaryote-dominated) metatranscriptomic analyses were conducted only on the larger pore-size filter (2.0 µm pore-size). All filters were immediately submerged in RNAlater (Applied Biosystems, Austin, TX) in sterile 50 mL conical tubes, incubated at room temperature overnight and then stored at -80oC until extraction. Filtration and stabilization of each sample was completed within 30 min of water collection.