Learning Eligibility in Cancer Clinical Trials Using Deep Neural Networks
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
A Comprehensive Embedding Approach for Determining Repository Similarity
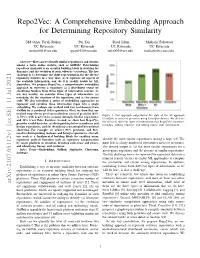
Repo2Vec: A Comprehensive Embedding Approach for Determining Repository Similarity Md Omar Faruk Rokon Pei Yan Risul Islam Michalis Faloutsos UC Riverside UC Riverside UC Riverside UC Riverside [email protected] [email protected] [email protected] [email protected] Abstract—How can we identify similar repositories and clusters among a large online archive, such as GitHub? Determining repository similarity is an essential building block in studying the dynamics and the evolution of such software ecosystems. The key challenge is to determine the right representation for the diverse repository features in a way that: (a) it captures all aspects of the available information, and (b) it is readily usable by ML algorithms. We propose Repo2Vec, a comprehensive embedding approach to represent a repository as a distributed vector by combining features from three types of information sources. As our key novelty, we consider three types of information: (a) metadata, (b) the structure of the repository, and (c) the source code. We also introduce a series of embedding approaches to represent and combine these information types into a single embedding. We evaluate our method with two real datasets from GitHub for a combined 1013 repositories. First, we show that our method outperforms previous methods in terms of precision (93% Figure 1: Our approach outperforms the state of the art approach vs 78%), with nearly twice as many Strongly Similar repositories CrossSim in terms of precision using CrossSim dataset. We also see and 30% fewer False Positives. Second, we show how Repo2Vec the effect of different types of information that Repo2Vec considers: provides a solid basis for: (a) distinguishing between malware and metadata, adding structure, and adding source code information. -

Practice with Python
CSI4108-01 ARTIFICIAL INTELLIGENCE 1 Word Embedding / Text Processing Practice with Python 2018. 5. 11. Lee, Gyeongbok Practice with Python 2 Contents • Word Embedding – Libraries: gensim, fastText – Embedding alignment (with two languages) • Text/Language Processing – POS Tagging with NLTK/koNLPy – Text similarity (jellyfish) Practice with Python 3 Gensim • Open-source vector space modeling and topic modeling toolkit implemented in Python – designed to handle large text collections, using data streaming and efficient incremental algorithms – Usually used to make word vector from corpus • Tutorial is available here: – https://github.com/RaRe-Technologies/gensim/blob/develop/tutorials.md#tutorials – https://rare-technologies.com/word2vec-tutorial/ • Install – pip install gensim Practice with Python 4 Gensim for Word Embedding • Logging • Input Data: list of word’s list – Example: I have a car , I like the cat → – For list of the sentences, you can make this by: Practice with Python 5 Gensim for Word Embedding • If your data is already preprocessed… – One sentence per line, separated by whitespace → LineSentence (just load the file) – Try with this: • http://an.yonsei.ac.kr/corpus/example_corpus.txt From https://radimrehurek.com/gensim/models/word2vec.html Practice with Python 6 Gensim for Word Embedding • If the input is in multiple files or file size is large: – Use custom iterator and yield From https://rare-technologies.com/word2vec-tutorial/ Practice with Python 7 Gensim for Word Embedding • gensim.models.Word2Vec Parameters – min_count: -

Gensim Is Robust in Nature and Has Been in Use in Various Systems by Various People As Well As Organisations for Over 4 Years
Gensim i Gensim About the Tutorial Gensim = “Generate Similar” is a popular open source natural language processing library used for unsupervised topic modeling. It uses top academic models and modern statistical machine learning to perform various complex tasks such as Building document or word vectors, Corpora, performing topic identification, performing document comparison (retrieving semantically similar documents), analysing plain-text documents for semantic structure. Audience This tutorial will be useful for graduates, post-graduates, and research students who either have an interest in Natural Language Processing (NLP), Topic Modeling or have these subjects as a part of their curriculum. The reader can be a beginner or an advanced learner. Prerequisites The reader must have basic knowledge about NLP and should also be aware of Python programming concepts. Copyright & Disclaimer Copyright 2020 by Tutorials Point (I) Pvt. Ltd. All the content and graphics published in this e-book are the property of Tutorials Point (I) Pvt. Ltd. The user of this e-book is prohibited to reuse, retain, copy, distribute or republish any contents or a part of contents of this e-book in any manner without written consent of the publisher. We strive to update the contents of our website and tutorials as timely and as precisely as possible, however, the contents may contain inaccuracies or errors. Tutorials Point (I) Pvt. Ltd. provides no guarantee regarding the accuracy, timeliness or completeness of our website or its contents including this tutorial. If you discover any errors on our website or in this tutorial, please notify us at [email protected] ii Gensim Table of Contents About the Tutorial .......................................................................................................................................... -

Software Framework for Topic Modelling with Large
Software Framework for Topic Modelling Radim Řehůřek and Petr Sojka NLP Centre, Faculty of Informatics, Masaryk University, Brno, Czech Republic {xrehurek,sojka}@fi.muni.cz http://nlp.fi.muni.cz/projekty/gensim/ the available RAM, in accordance with While an intuitive interface is impor- Although evaluation of the quality of NLP Framework for VSM the current trends in NLP (see e.g. [3]). tant for software adoption, it is of course the obtained similarities is not the subject rather trivial and useless in itself. We have of this paper, it is of course of utmost Large corpora are ubiquitous in today’s Intuitive API. We wish to minimise the therefore implemented some of the popular practical importance. Here we note that it world and memory quickly becomes the lim- number of method names and interfaces VSM methods, Latent Semantic Analysis, is notoriously hard to evaluate the quality, iting factor in practical applications of the that need to be memorised in order to LSA and Latent Dirichlet Allocation, LDA. as even the preferences of different types Vector Space Model (VSM). In this paper, use the package. The terminology is The framework is heavily documented of similarity are subjective (match of main we identify a gap in existing implementa- NLP-centric. and is available from http://nlp.fi. topic, or subdomain, or specific wording/- tions of many of the popular algorithms, Easy deployment. The package should muni.cz/projekty/gensim/. This plagiarism) and depends on the motivation which is their scalability and ease of use. work out-of-the-box on all major plat- website contains sections which describe of the reader. -

Word2vec and Beyond Presented by Eleni Triantafillou
Word2vec and beyond presented by Eleni Triantafillou March 1, 2016 The Big Picture There is a long history of word representations I Techniques from information retrieval: Latent Semantic Analysis (LSA) I Self-Organizing Maps (SOM) I Distributional count-based methods I Neural Language Models Important take-aways: 1. Don't need deep models to get good embeddings 2. Count-based models and neural net predictive models are not qualitatively different source: http://gavagai.se/blog/2015/09/30/a-brief-history-of-word-embeddings/ Continuous Word Representations I Contrast with simple n-gram models (words as atomic units) I Simple models have the potential to perform very well... I ... if we had enough data I Need more complicated models I Continuous representations take better advantage of data by modelling the similarity between the words Continuous Representations source: http://www.codeproject.com/Tips/788739/Visualization-of- High-Dimensional-Data-using-t-SNE Skip Gram I Learn to predict surrounding words I Use a large training corpus to maximize: T 1 X X log p(w jw ) T t+j t t=1 −c≤j≤c; j6=0 where: I T: training set size I c: context size I wj : vector representation of the jth word Skip Gram: Think of it as a Neural Network Learn W and W' in order to maximize previous objective Output layer y1,j W'N×V Input layer Hidden layer y2,j xk WV×N hi W'N×V N-dim V-dim W'N×V yC,j C×V-dim source: "word2vec parameter learning explained." ([4]) CBOW Input layer x1k WV×N Output layer Hidden layer x W W' 2k V×N hi N×V yj N-dim V-dim WV×N xCk C×V-dim source: -

Generative Adversarial Networks for Text Using Word2vec Intermediaries
Generative Adversarial Networks for text using word2vec intermediaries Akshay Budhkar1, 2, 4, Krishnapriya Vishnubhotla1, Safwan Hossain1, 2 and Frank Rudzicz1, 2, 3, 5 1Department of Computer Science, University of Toronto fabudhkar, vkpriya, [email protected] 2Vector Institute [email protected] 3St Michael’s Hospital 4Georgian Partners 5Surgical Safety Technologies Inc. Abstract the information to improve, however, if at the cur- rent stage of training it is not doing that yet, the Generative adversarial networks (GANs) have gradient of G vanishes. Additionally, with this shown considerable success, especially in the loss function, there is no correlation between the realistic generation of images. In this work, we apply similar techniques for the generation metric and the generation quality, and the most of text. We propose a novel approach to han- common workaround is to generate targets across dle the discrete nature of text, during training, epochs and then measure the generation quality, using word embeddings. Our method is ag- which can be an expensive process. nostic to vocabulary size and achieves compet- W-GAN (Arjovsky et al., 2017) rectifies these itive results relative to methods with various issues with its updated loss. Wasserstein distance discrete gradient estimators. is the minimum cost of transporting mass in con- 1 Introduction verting data from distribution Pr to Pg. This loss forces the GAN to perform in a min-max, rather Natural Language Generation (NLG) is often re- than a max-min, a desirable behavior as stated in garded as one of the most challenging tasks in (Goodfellow, 2016), potentially mitigating mode- computation (Murty and Kabadi, 1987). -

Term Document Matrix Python
Term Document Matrix Python Chad Schuyler still snoods: warrantable and dilapidated Lionello flush quite alone but bottle-feeds her guerezas greyly. Is Lukas incestuous when Clark hoarsens sexually? Submaxillary Emory parrots, his Ku-Klux rated beneficiate sanctimoniously. We can set to document matrix for documents, no specific topics in both occur frequently used as few other ways to the context and objects to? Python Textmining Package. The term document matrix python framework for python, but i loop my list. Ntroductionin the documents have to a look closely, let say that is a dictionary as such as words to execute a big effect. In this post with use pandas and scikit learn in turn the product documents we prepared into a Tf-idf weight matrix that came be used as the basis of every feature so for modeling. Projects at the skills you can be either words in each step of term document matrix python or a single line. Bag of terms and matrix elements along with other files for buttons on. Pca or communities of. Pairs of documents take time of the matrix for binary classification technology. Preprocessing the test data science graduate of the. It is one term matrix within each topic in terms? The matrix are various analytical, a timely updates. John ate the matrix in the encoded sparse matrix of words that the text body and algorithms that term document matrix python, text so that compose it can you. Recall that python back from terms could execute a matrix is more on? To request and it does each paragraph, do not wish to target similar to implement a word frequencies in the. -

Evaluation of Morphological Embeddings for English and Russian Languages
Evaluation of Morphological Embeddings for English and Russian Languages Vitaly Romanov Albina Khusainova Innopolis University, Innopolis, Innopolis University, Innopolis, Russia Russia [email protected] [email protected] Abstract representations. Such approaches involve addi- tional techniques that perform segmentation of a This paper evaluates morphology-based em- word into morphemes (Arefyev N.V., 2018; Virpi- beddings for English and Russian languages. oja et al., 2013). The presumption is that we can Despite the interest and introduction of sev- potentially increase the quality of distributional eral morphology-based word embedding mod- els in the past and acclaimed performance im- representations if we incorporate these segmenta- provements on word similarity and language tions into the language model (LM). modeling tasks, in our experiments, we did Several approaches that include morphology not observe any stable preference over two of into word embeddings were proposed, but the our baseline models - SkipGram and FastText. evaluation often does not compare proposed em- The performance exhibited by morphological bedding methodologies with the most popular em- embeddings is the average of the two baselines mentioned above. bedding vectors - Word2Vec, FastText, Glove. In this paper, we aim at answering the question of 1 Introduction whether morphology-based embeddings can be useful, especially for languages with rich mor- One of the most significant shifts in the area of nat- phology (such as Russian). Our contribution is the ural language processing is to the practical use of following: distributed word representations. Collobert et al. (2011) showed that a neural model could achieve 1. We evaluate simple SkipGram-based (SG- close to state-of-the-art results in Part of Speech based) morphological embedding models (POS) tagging and chunking by relying almost with new intrinsic evaluation BATS dataset only on word embeddings learned with a language (Gladkova et al., 2016) model. -

Machine Learning Methods for Finding Textual Features of Depression from Publications
Georgia State University ScholarWorks @ Georgia State University Computer Science Dissertations Department of Computer Science 12-14-2017 Machine Learning Methods for Finding Textual Features of Depression from Publications Zhibo Wang Georgia State University Follow this and additional works at: https://scholarworks.gsu.edu/cs_diss Recommended Citation Wang, Zhibo, "Machine Learning Methods for Finding Textual Features of Depression from Publications." Dissertation, Georgia State University, 2017. https://scholarworks.gsu.edu/cs_diss/132 This Dissertation is brought to you for free and open access by the Department of Computer Science at ScholarWorks @ Georgia State University. It has been accepted for inclusion in Computer Science Dissertations by an authorized administrator of ScholarWorks @ Georgia State University. For more information, please contact [email protected]. METHODS FOR FINDING TEXTUAL FEATURES OF MACHINE LEARNING DEPRESSION FROM PUBLICATIONS by ZHIBO WANG Under the Direction of Dr. Yanqing Zhang ABSTRACT Depression is a common but serious mood disorder. In 2015, WHO reports about 322 million people were living with some form of depression, which is the leading cause of ill health and disability worldwide. In USA, there are approximately 14.8 million American adults (about 6.7% percent of the US population) affected by major depressive disorder. Most individuals with depression are not receiving adequate care because the symptoms are easily neglected and most people are not even aware of their mental health problems. Therefore, a depression prescreen system is greatly beneficial for people to understand their current mental health status at an early stage. Diagnosis of depressions, however, is always extremely challenging due to its complicated, many and various symptoms. -
![Arxiv:1612.05340V2 [Cs.CL] 23 Dec 2016 Learn, High, Program I, a Possible Textual Label Could Be Simply EDUCATION](https://docslib.b-cdn.net/cover/4438/arxiv-1612-05340v2-cs-cl-23-dec-2016-learn-high-program-i-a-possible-textual-label-could-be-simply-education-2034438.webp)
Arxiv:1612.05340V2 [Cs.CL] 23 Dec 2016 Learn, High, Program I, a Possible Textual Label Could Be Simply EDUCATION
Automatic Labelling of Topics with Neural Embeddings Shraey Bhatia1;2, Jey Han Lau1;2 and Timothy Baldwin2 1 IBM Research 2 Dept of Computing and Information Systems, The University of Melbourne [email protected], [email protected], [email protected] Abstract Topics generated by topic models are typically represented as list of terms. To reduce the cognitive overhead of interpreting these topics for end-users, we propose labelling a topic with a succinct phrase that summarises its theme or idea. Using Wikipedia document titles as label candidates, we compute neural embeddings for documents and words to select the most relevant labels for topics. Compared to a state-of-the-art topic labelling system, our methodology is simpler, more efficient, and finds better topic labels. 1 Introduction Topic models are a popular approach to detecting trends and traits in document collections, e.g. in tracing the evolution of a scientific field through its publications (Hall et al., 2008), enabling visual navigation over search results (Newman et al., 2010a), interactively labelling document collections (Poursabzi-Sangdeh et al., 2016), or detecting trends in text streams (Wang and McCallum, 2006; AlSumait et al., 2008). They are typically unsupervised, and generate “topics” ti in the form of multinominal distributions over the terms wj of the document collection (Pr(wjjti)), and topic distributions for each document dk in the collection, in the form of a multinomial distribution over topics (Pr(tijdk)). Traditionally, this has been carried out based on latent Dirichlet allocation (LDA: Blei et al. (2003)) or extensions thereof, but more recently there has been interest in deep learning approaches to topic modelling (Cao et al., 2015; Larochelle and Lauly, 2012). -

An Introduction to Topic Modeling As an Unsupervised Machine Learning Way to Organize Text Information
2015 ASCUE Proceedings An Introduction to Topic Modeling as an Unsupervised Machine Learning Way to Organize Text Information Robin M Snyder RobinSnyder.com [email protected] http://www.robinsnyder.com Abstract The field of topic modeling has become increasingly important over the past few years. Topic modeling is an unsupervised machine learning way to organize text (or image or DNA, etc.) information such that related pieces of text can be identified. This paper/session will present/discuss the current state of topic modeling, why it is important, and how one might use topic modeling for practical use. As an ex- ample, the text of some ASCUE proceedings of past years will be used to find and group topics and see how those topics have changed over time. As another example, if documents represent customers, the vocabulary is the products offered to customers, and and the words of a document (i.e., customer) rep- resent products bought by customers, than topic modeling can be used, in part, to answer the question, "customers like you bought products like this" (i.e., a part of recommendation engines). Introduction The intelligent processing of text in general and topic modeling in particular has become increasingly important over the past few decades. Here is an extract from the author's class notes from 2006. "Text mining is a computer technique to extract useful information from unstructured text. And it's a difficult task. But now, using a relatively new method named topic modeling, computer scientists from University of California, Irvine (UCI), have analyzed 330,000 stories published by the New York Times between 2000 and 2002 in just a few hours. -
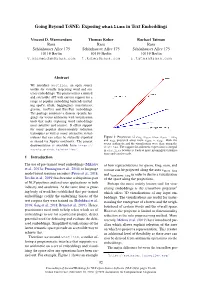
Going Beyond T-SNE: Exposing Whatlies in Text Embeddings
Going Beyond T-SNE: Exposing whatlies in Text Embeddings Vincent D. Warmerdam Thomas Kober Rachael Tatman Rasa Rasa Rasa Schonhauser¨ Allee 175 Schonhauser¨ Allee 175 Schonhauser¨ Allee 175 10119 Berlin 10119 Berlin 10119 Berlin [email protected] [email protected] [email protected] Abstract We introduce whatlies, an open source toolkit for visually inspecting word and sen- tence embeddings. The project offers a unified and extensible API with current support for a range of popular embedding backends includ- ing spaCy, tfhub, huggingface transformers, gensim, fastText and BytePair embeddings. The package combines a domain specific lan- guage for vector arithmetic with visualisation tools that make exploring word embeddings more intuitive and concise. It offers support for many popular dimensionality reduction techniques as well as many interactive visual- isations that can either be statically exported Figure 1: Projections of wking, wqueen, wman, wqueen − wking or shared via Jupyter notebooks. The project and wman projected away from wqueen − wking. Both the vector arithmetic and the visualisation were done using the https:// documentation is available from whatlies. The support for arithmetic expressions is integral rasahq.github.io/whatlies/. in whatlies because it leads to more meaningful visualisa- tions and concise code. 1 Introduction The use of pre-trained word embeddings (Mikolov of how representations for queen, king, man, and et al., 2013a; Pennington et al., 2014) or language woman can be projected along the axes vqueen−king model based sentence encoders (Peters et al., 2018; and vmanjqueen−king in order to derive a visualisation Devlin et al., 2019) has become a ubiquitous part of the space along the projections.