Gensim Documentation Release 0.6.0
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
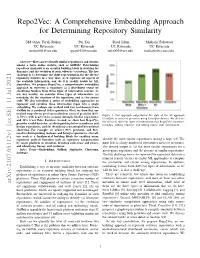
A Comprehensive Embedding Approach for Determining Repository Similarity
Repo2Vec: A Comprehensive Embedding Approach for Determining Repository Similarity Md Omar Faruk Rokon Pei Yan Risul Islam Michalis Faloutsos UC Riverside UC Riverside UC Riverside UC Riverside [email protected] [email protected] [email protected] [email protected] Abstract—How can we identify similar repositories and clusters among a large online archive, such as GitHub? Determining repository similarity is an essential building block in studying the dynamics and the evolution of such software ecosystems. The key challenge is to determine the right representation for the diverse repository features in a way that: (a) it captures all aspects of the available information, and (b) it is readily usable by ML algorithms. We propose Repo2Vec, a comprehensive embedding approach to represent a repository as a distributed vector by combining features from three types of information sources. As our key novelty, we consider three types of information: (a) metadata, (b) the structure of the repository, and (c) the source code. We also introduce a series of embedding approaches to represent and combine these information types into a single embedding. We evaluate our method with two real datasets from GitHub for a combined 1013 repositories. First, we show that our method outperforms previous methods in terms of precision (93% Figure 1: Our approach outperforms the state of the art approach vs 78%), with nearly twice as many Strongly Similar repositories CrossSim in terms of precision using CrossSim dataset. We also see and 30% fewer False Positives. Second, we show how Repo2Vec the effect of different types of information that Repo2Vec considers: provides a solid basis for: (a) distinguishing between malware and metadata, adding structure, and adding source code information. -

Practice with Python
CSI4108-01 ARTIFICIAL INTELLIGENCE 1 Word Embedding / Text Processing Practice with Python 2018. 5. 11. Lee, Gyeongbok Practice with Python 2 Contents • Word Embedding – Libraries: gensim, fastText – Embedding alignment (with two languages) • Text/Language Processing – POS Tagging with NLTK/koNLPy – Text similarity (jellyfish) Practice with Python 3 Gensim • Open-source vector space modeling and topic modeling toolkit implemented in Python – designed to handle large text collections, using data streaming and efficient incremental algorithms – Usually used to make word vector from corpus • Tutorial is available here: – https://github.com/RaRe-Technologies/gensim/blob/develop/tutorials.md#tutorials – https://rare-technologies.com/word2vec-tutorial/ • Install – pip install gensim Practice with Python 4 Gensim for Word Embedding • Logging • Input Data: list of word’s list – Example: I have a car , I like the cat → – For list of the sentences, you can make this by: Practice with Python 5 Gensim for Word Embedding • If your data is already preprocessed… – One sentence per line, separated by whitespace → LineSentence (just load the file) – Try with this: • http://an.yonsei.ac.kr/corpus/example_corpus.txt From https://radimrehurek.com/gensim/models/word2vec.html Practice with Python 6 Gensim for Word Embedding • If the input is in multiple files or file size is large: – Use custom iterator and yield From https://rare-technologies.com/word2vec-tutorial/ Practice with Python 7 Gensim for Word Embedding • gensim.models.Word2Vec Parameters – min_count: -

Document and Topic Models: Plsa
10/4/2018 Document and Topic Models: pLSA and LDA Andrew Levandoski and Jonathan Lobo CS 3750 Advanced Topics in Machine Learning 2 October 2018 Outline • Topic Models • pLSA • LSA • Model • Fitting via EM • pHITS: link analysis • LDA • Dirichlet distribution • Generative process • Model • Geometric Interpretation • Inference 2 1 10/4/2018 Topic Models: Visual Representation Topic proportions and Topics Documents assignments 3 Topic Models: Importance • For a given corpus, we learn two things: 1. Topic: from full vocabulary set, we learn important subsets 2. Topic proportion: we learn what each document is about • This can be viewed as a form of dimensionality reduction • From large vocabulary set, extract basis vectors (topics) • Represent document in topic space (topic proportions) 푁 퐾 • Dimensionality is reduced from 푤푖 ∈ ℤ푉 to 휃 ∈ ℝ • Topic proportion is useful for several applications including document classification, discovery of semantic structures, sentiment analysis, object localization in images, etc. 4 2 10/4/2018 Topic Models: Terminology • Document Model • Word: element in a vocabulary set • Document: collection of words • Corpus: collection of documents • Topic Model • Topic: collection of words (subset of vocabulary) • Document is represented by (latent) mixture of topics • 푝 푤 푑 = 푝 푤 푧 푝(푧|푑) (푧 : topic) • Note: document is a collection of words (not a sequence) • ‘Bag of words’ assumption • In probability, we call this the exchangeability assumption • 푝 푤1, … , 푤푁 = 푝(푤휎 1 , … , 푤휎 푁 ) (휎: permutation) 5 Topic Models: Terminology (cont’d) • Represent each document as a vector space • A word is an item from a vocabulary indexed by {1, … , 푉}. We represent words using unit‐basis vectors. -

Redalyc.Latent Dirichlet Allocation Complement in the Vector Space Model for Multi-Label Text Classification
International Journal of Combinatorial Optimization Problems and Informatics E-ISSN: 2007-1558 [email protected] International Journal of Combinatorial Optimization Problems and Informatics México Carrera-Trejo, Víctor; Sidorov, Grigori; Miranda-Jiménez, Sabino; Moreno Ibarra, Marco; Cadena Martínez, Rodrigo Latent Dirichlet Allocation complement in the vector space model for Multi-Label Text Classification International Journal of Combinatorial Optimization Problems and Informatics, vol. 6, núm. 1, enero-abril, 2015, pp. 7-19 International Journal of Combinatorial Optimization Problems and Informatics Morelos, México Available in: http://www.redalyc.org/articulo.oa?id=265239212002 How to cite Complete issue Scientific Information System More information about this article Network of Scientific Journals from Latin America, the Caribbean, Spain and Portugal Journal's homepage in redalyc.org Non-profit academic project, developed under the open access initiative © International Journal of Combinatorial Optimization Problems and Informatics, Vol. 6, No. 1, Jan-April 2015, pp. 7-19. ISSN: 2007-1558. Latent Dirichlet Allocation complement in the vector space model for Multi-Label Text Classification Víctor Carrera-Trejo1, Grigori Sidorov1, Sabino Miranda-Jiménez2, Marco Moreno Ibarra1 and Rodrigo Cadena Martínez3 Centro de Investigación en Computación1, Instituto Politécnico Nacional, México DF, México Centro de Investigación e Innovación en Tecnologías de la Información y Comunicación (INFOTEC)2, Ags., México Universidad Tecnológica de México3 – UNITEC MÉXICO [email protected], {sidorov,marcomoreno}@cic.ipn.mx, [email protected] [email protected] Abstract. In text classification task one of the main problems is to choose which features give the best results. Various features can be used like words, n-grams, syntactic n-grams of various types (POS tags, dependency relations, mixed, etc.), or a combinations of these features can be considered. -

Evaluating Vector-Space Models of Word Representation, Or, the Unreasonable Effectiveness of Counting Words Near Other Words
Evaluating Vector-Space Models of Word Representation, or, The Unreasonable Effectiveness of Counting Words Near Other Words Aida Nematzadeh, Stephan C. Meylan, and Thomas L. Griffiths University of California, Berkeley fnematzadeh, smeylan, tom griffi[email protected] Abstract angle between word vectors (e.g., Mikolov et al., 2013b; Pen- nington et al., 2014). Vector-space models of semantics represent words as continuously-valued vectors and measure similarity based on In this paper, we examine whether these constraints im- the distance or angle between those vectors. Such representa- ply that Word2Vec and GloVe representations suffer from the tions have become increasingly popular due to the recent de- same difficulty as previous vector-space models in capturing velopment of methods that allow them to be efficiently esti- mated from very large amounts of data. However, the idea human similarity judgments. To this end, we evaluate these of relating similarity to distance in a spatial representation representations on a set of tasks adopted from Griffiths et al. has been criticized by cognitive scientists, as human similar- (2007) in which the authors showed that the representations ity judgments have many properties that are inconsistent with the geometric constraints that a distance metric must obey. We learned by another well-known vector-space model, Latent show that two popular vector-space models, Word2Vec and Semantic Analysis (Landauer and Dumais, 1997), were in- GloVe, are unable to capture certain critical aspects of human consistent with patterns of semantic similarity demonstrated word association data as a consequence of these constraints. However, a probabilistic topic model estimated from a rela- in human word association data. -

Gensim Is Robust in Nature and Has Been in Use in Various Systems by Various People As Well As Organisations for Over 4 Years
Gensim i Gensim About the Tutorial Gensim = “Generate Similar” is a popular open source natural language processing library used for unsupervised topic modeling. It uses top academic models and modern statistical machine learning to perform various complex tasks such as Building document or word vectors, Corpora, performing topic identification, performing document comparison (retrieving semantically similar documents), analysing plain-text documents for semantic structure. Audience This tutorial will be useful for graduates, post-graduates, and research students who either have an interest in Natural Language Processing (NLP), Topic Modeling or have these subjects as a part of their curriculum. The reader can be a beginner or an advanced learner. Prerequisites The reader must have basic knowledge about NLP and should also be aware of Python programming concepts. Copyright & Disclaimer Copyright 2020 by Tutorials Point (I) Pvt. Ltd. All the content and graphics published in this e-book are the property of Tutorials Point (I) Pvt. Ltd. The user of this e-book is prohibited to reuse, retain, copy, distribute or republish any contents or a part of contents of this e-book in any manner without written consent of the publisher. We strive to update the contents of our website and tutorials as timely and as precisely as possible, however, the contents may contain inaccuracies or errors. Tutorials Point (I) Pvt. Ltd. provides no guarantee regarding the accuracy, timeliness or completeness of our website or its contents including this tutorial. If you discover any errors on our website or in this tutorial, please notify us at [email protected] ii Gensim Table of Contents About the Tutorial .......................................................................................................................................... -

Software Framework for Topic Modelling with Large
Software Framework for Topic Modelling Radim Řehůřek and Petr Sojka NLP Centre, Faculty of Informatics, Masaryk University, Brno, Czech Republic {xrehurek,sojka}@fi.muni.cz http://nlp.fi.muni.cz/projekty/gensim/ the available RAM, in accordance with While an intuitive interface is impor- Although evaluation of the quality of NLP Framework for VSM the current trends in NLP (see e.g. [3]). tant for software adoption, it is of course the obtained similarities is not the subject rather trivial and useless in itself. We have of this paper, it is of course of utmost Large corpora are ubiquitous in today’s Intuitive API. We wish to minimise the therefore implemented some of the popular practical importance. Here we note that it world and memory quickly becomes the lim- number of method names and interfaces VSM methods, Latent Semantic Analysis, is notoriously hard to evaluate the quality, iting factor in practical applications of the that need to be memorised in order to LSA and Latent Dirichlet Allocation, LDA. as even the preferences of different types Vector Space Model (VSM). In this paper, use the package. The terminology is The framework is heavily documented of similarity are subjective (match of main we identify a gap in existing implementa- NLP-centric. and is available from http://nlp.fi. topic, or subdomain, or specific wording/- tions of many of the popular algorithms, Easy deployment. The package should muni.cz/projekty/gensim/. This plagiarism) and depends on the motivation which is their scalability and ease of use. work out-of-the-box on all major plat- website contains sections which describe of the reader. -

Deconstructing Word Embedding Models
Deconstructing Word Embedding Models Koushik K. Varma Ashoka University, [email protected] Abstract – Analysis of Word Embedding Models through Analysis, one must be certain that all linguistic aspects of a a deconstructive approach reveals their several word are captured within the vector representations because shortcomings and inconsistencies. These include any discrepancies could be further amplified in practical instability of the vector representations, a distorted applications. While these vector representations have analogical reasoning, geometric incompatibility with widespread use in modern natural language processing, it is linguistic features, and the inconsistencies in the corpus unclear as to what degree they accurately encode the data. A new theoretical embedding model, ‘Derridian essence of language in its structural, contextual, cultural and Embedding,’ is proposed in this paper. Contemporary ethical assimilation. Surface evaluations on training-test embedding models are evaluated qualitatively in terms data provide efficiency measures for a specific of how adequate they are in relation to the capabilities of implementation but do not explicitly give insights on the a Derridian Embedding. factors causing the existing efficiency (or the lack there of). We henceforth present a deconstructive review of 1. INTRODUCTION contemporary word embeddings using available literature to clarify certain misconceptions and inconsistencies Saussure, in the Course in General Linguistics, concerning them. argued that there is no substance in language and that all language consists of differences. In language there are only forms, not substances, and by that he meant that all 1.1 DECONSTRUCTIVE APPROACH apparently substantive units of language are generated by other things that lie outside them, but these external Derrida, a post-structualist French philosopher, characteristics are actually internal to their make-up. -

Word2vec and Beyond Presented by Eleni Triantafillou
Word2vec and beyond presented by Eleni Triantafillou March 1, 2016 The Big Picture There is a long history of word representations I Techniques from information retrieval: Latent Semantic Analysis (LSA) I Self-Organizing Maps (SOM) I Distributional count-based methods I Neural Language Models Important take-aways: 1. Don't need deep models to get good embeddings 2. Count-based models and neural net predictive models are not qualitatively different source: http://gavagai.se/blog/2015/09/30/a-brief-history-of-word-embeddings/ Continuous Word Representations I Contrast with simple n-gram models (words as atomic units) I Simple models have the potential to perform very well... I ... if we had enough data I Need more complicated models I Continuous representations take better advantage of data by modelling the similarity between the words Continuous Representations source: http://www.codeproject.com/Tips/788739/Visualization-of- High-Dimensional-Data-using-t-SNE Skip Gram I Learn to predict surrounding words I Use a large training corpus to maximize: T 1 X X log p(w jw ) T t+j t t=1 −c≤j≤c; j6=0 where: I T: training set size I c: context size I wj : vector representation of the jth word Skip Gram: Think of it as a Neural Network Learn W and W' in order to maximize previous objective Output layer y1,j W'N×V Input layer Hidden layer y2,j xk WV×N hi W'N×V N-dim V-dim W'N×V yC,j C×V-dim source: "word2vec parameter learning explained." ([4]) CBOW Input layer x1k WV×N Output layer Hidden layer x W W' 2k V×N hi N×V yj N-dim V-dim WV×N xCk C×V-dim source: -

Generative Adversarial Networks for Text Using Word2vec Intermediaries
Generative Adversarial Networks for text using word2vec intermediaries Akshay Budhkar1, 2, 4, Krishnapriya Vishnubhotla1, Safwan Hossain1, 2 and Frank Rudzicz1, 2, 3, 5 1Department of Computer Science, University of Toronto fabudhkar, vkpriya, [email protected] 2Vector Institute [email protected] 3St Michael’s Hospital 4Georgian Partners 5Surgical Safety Technologies Inc. Abstract the information to improve, however, if at the cur- rent stage of training it is not doing that yet, the Generative adversarial networks (GANs) have gradient of G vanishes. Additionally, with this shown considerable success, especially in the loss function, there is no correlation between the realistic generation of images. In this work, we apply similar techniques for the generation metric and the generation quality, and the most of text. We propose a novel approach to han- common workaround is to generate targets across dle the discrete nature of text, during training, epochs and then measure the generation quality, using word embeddings. Our method is ag- which can be an expensive process. nostic to vocabulary size and achieves compet- W-GAN (Arjovsky et al., 2017) rectifies these itive results relative to methods with various issues with its updated loss. Wasserstein distance discrete gradient estimators. is the minimum cost of transporting mass in con- 1 Introduction verting data from distribution Pr to Pg. This loss forces the GAN to perform in a min-max, rather Natural Language Generation (NLG) is often re- than a max-min, a desirable behavior as stated in garded as one of the most challenging tasks in (Goodfellow, 2016), potentially mitigating mode- computation (Murty and Kabadi, 1987). -

Vector Space Models for Words and Documents Statistical Natural Language Processing ELEC-E5550 Jan 30, 2019 Tiina Lindh-Knuutila D.Sc
Vector space models for words and documents Statistical Natural Language Processing ELEC-E5550 Jan 30, 2019 Tiina Lindh-Knuutila D.Sc. (Tech) Lingsoft Language Services & Aalto University tiina.lindh-knuutila -at- aalto dot fi Material also by Mari-Sanna Paukkeri (mari-sanna.paukkeri -at- utopiaanalytics dot com) Today’s Agenda • Vector space models • word-document matrices • word vectors • stemming, weighting, dimensionality reduction • similarity measures • Count models vs. predictive models • Word2vec • Information retrieval (Briefly) • Course project details You shall know the word by the company it keeps • Language is symbolic in nature • Surface form is in an arbitrary relation with the meaning of the word • Hat vs. cat • One substitution: Levenshtein distance of 1 • Does not measure the semantic similarity of the words • Distributional semantics • Linguistic items which appear in similar contexts in large samples of language data tend to have similar meanings Firth, John R. 1957. A synopsis of linguistic theory 1930–1955. In Studies in linguistic analysis, 1–32. Oxford: Blackwell. George Miller and Walter Charles. 1991. Contextual correlates of semantic similarity. Language and Cognitive Processes, 6(1):1–28. Vector space models (VSM) • The use of a high-dimensional space of documents (or words) • Closeness in the vector space resembles closeness in the semantics or structure of the documents (depending on the features extracted). • Makes the use of data mining possible • Applications: – Document clustering/classification/… • Finding similar documents • Finding similar words – Word disambiguation – Information retrieval • Term discrimination: ranking keywords in the order of usefulness Vector space models (VSM) • Steps to build a vector space model 1. Preprocessing 2. -

Word Embeddings: History & Behind the Scene
Word Embeddings: History & Behind the Scene Alfan F. Wicaksono Information Retrieval Lab. Faculty of Computer Science Universitas Indonesia References • Bengio, Y., Ducharme, R., Vincent, P., & Janvin, C. (2003). A Neural Probabilistic Language Model. The Journal of Machine Learning Research, 3, 1137–1155. • Mikolov, T., Corrado, G., Chen, K., & Dean, J. (2013). Efficient Estimation of Word Representations in Vector Space. Proceedings of the International Conference on Learning Representations (ICLR 2013), 1–12 • Mikolov, T., Chen, K., Corrado, G., & Dean, J. (2013). Distributed Representations of Words and Phrases and their Compositionality. NIPS, 1–9. • Morin, F., & Bengio, Y. (2005). Hierarchical Probabilistic Neural Network Language Model. Aistats, 5. References Good weblogs for high-level understanding: • http://sebastianruder.com/word-embeddings-1/ • Sebastian Ruder. On word embeddings - Part 2: Approximating the Softmax. http://sebastianruder.com/word- embeddings-softmax • https://www.tensorflow.org/tutorials/word2vec • https://www.gavagai.se/blog/2015/09/30/a-brief-history-of- word-embeddings/ Some slides were also borrowed from From Dan Jurafsky’s course slide: Word Meaning and Similarity. Stanford University. Terminology • The term “Word Embedding” came from deep learning community • For computational linguistic community, they prefer “Distributional Semantic Model” • Other terms: – Distributed Representation – Semantic Vector Space – Word Space https://www.gavagai.se/blog/2015/09/30/a-brief-history-of-word-embeddings/ Before we learn Word Embeddings... Semantic Similarity • Word Similarity – Near-synonyms – “boat” and “ship”, “car” and “bicycle” • Word Relatedness – Can be related any way – Similar: “boat” and “ship” – Topical Similarity: • “boat” and “water” • “car” and “gasoline” Why Word Similarity? • Document Classification • Document Clustering • Language Modeling • Information Retrieval • ..