The Ploidy Races of Atriplex Confertifolia (Chenopodiaceae)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

California Vegetation Map in Support of the DRECP
CALIFORNIA VEGETATION MAP IN SUPPORT OF THE DESERT RENEWABLE ENERGY CONSERVATION PLAN (2014-2016 ADDITIONS) John Menke, Edward Reyes, Anne Hepburn, Deborah Johnson, and Janet Reyes Aerial Information Systems, Inc. Prepared for the California Department of Fish and Wildlife Renewable Energy Program and the California Energy Commission Final Report May 2016 Prepared by: Primary Authors John Menke Edward Reyes Anne Hepburn Deborah Johnson Janet Reyes Report Graphics Ben Johnson Cover Page Photo Credits: Joshua Tree: John Fulton Blue Palo Verde: Ed Reyes Mojave Yucca: John Fulton Kingston Range, Pinyon: Arin Glass Aerial Information Systems, Inc. 112 First Street Redlands, CA 92373 (909) 793-9493 [email protected] in collaboration with California Department of Fish and Wildlife Vegetation Classification and Mapping Program 1807 13th Street, Suite 202 Sacramento, CA 95811 and California Native Plant Society 2707 K Street, Suite 1 Sacramento, CA 95816 i ACKNOWLEDGEMENTS Funding for this project was provided by: California Energy Commission US Bureau of Land Management California Wildlife Conservation Board California Department of Fish and Wildlife Personnel involved in developing the methodology and implementing this project included: Aerial Information Systems: Lisa Cotterman, Mark Fox, John Fulton, Arin Glass, Anne Hepburn, Ben Johnson, Debbie Johnson, John Menke, Lisa Morse, Mike Nelson, Ed Reyes, Janet Reyes, Patrick Yiu California Department of Fish and Wildlife: Diana Hickson, Todd Keeler‐Wolf, Anne Klein, Aicha Ougzin, Rosalie Yacoub California -

Appendix F3 Rare Plant Survey Report
Appendix F3 Rare Plant Survey Report Draft CADIZ VALLEY WATER CONSERVATION, RECOVERY, AND STORAGE PROJECT Rare Plant Survey Report Prepared for May 2011 Santa Margarita Water District Draft CADIZ VALLEY WATER CONSERVATION, RECOVERY, AND STORAGE PROJECT Rare Plant Survey Report Prepared for May 2011 Santa Margarita Water District 626 Wilshire Boulevard Suite 1100 Los Angeles, CA 90017 213.599.4300 www.esassoc.com Oakland Olympia Petaluma Portland Sacramento San Diego San Francisco Seattle Tampa Woodland Hills D210324 TABLE OF CONTENTS Cadiz Valley Water Conservation, Recovery, and Storage Project: Rare Plant Survey Report Page Summary ............................................................................................................................... 1 Introduction ..........................................................................................................................2 Objective .......................................................................................................................... 2 Project Location and Description .....................................................................................2 Setting ................................................................................................................................... 5 Climate ............................................................................................................................. 5 Topography and Soils ......................................................................................................5 -

Foraoe Plants for ( Fltiyation on Alkali Soils
FORAOE PLANTS FOR ( FLTIYATION ON ALKALI SOILS. By JARED G. SMITH, Assistant Agrostologist. INTRODUCTION. When the excess of alkali in a soil rises to such a sum total that the cereals can not be grown, the best use to which the land can be put is the growth of forage. There is no crop, so far as known, which can be profitably grown on land that contains over 2 per cent of the combined alkalies within the first 2 inches of surface soil. Soils containing as much as 1 per cent of alkalies within the first 2 inches will not grow cereals, and the maximum for trees, vines, and root crops is much lower. There are extensive areas in the West which are thus excluded from the cate- gory of farming lands because of the excess of alkalies. Such lands are not necessarily barren, although they can not be profitably culti- vated. They are often covered by a rank growth of vegetation, indi- cating that there is an abundance of plant food in the soil, if only the plants are so tolerant of alkali as to be able to secure it. Many of the alkaii plants have considerable value as forage, and of these some few show special adaptation to the changed conditions which cultiva- tion brings. The crops which originated in humid regions will not grow on soils which are strongly impregnated with the alkalies, and so to meet the conditions one must either take the useful plants of the alkali regions of his own country or depend upon those which have been introduced from similar regions elsewhere. -
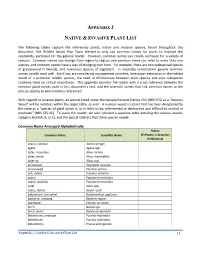
Reference Plant List
APPENDIX J NATIVE & INVASIVE PLANT LIST The following tables capture the referenced plants, native and invasive species, found throughout this document. The Wildlife Action Plan Team elected to only use common names for plants to improve the readability, particular for the general reader. However, common names can create confusion for a variety of reasons. Common names can change from region-to-region; one common name can refer to more than one species; and common names have a way of changing over time. For example, there are two widespread species of greasewood in Nevada, and numerous species of sagebrush. In everyday conversation generic common names usually work well. But if you are considering management activities, landscape restoration or the habitat needs of a particular wildlife species, the need to differentiate between plant species and even subspecies suddenly takes on critical importance. This appendix provides the reader with a cross reference between the common plant names used in this document’s text, and the scientific names that link common names to the precise species to which writers referenced. With regards to invasive plants, all species listed under the Nevada Revised Statute 555 (NRS 555) as a “Noxious Weed” will be notated, within the larger table, as such. A noxious weed is a plant that has been designated by the state as a “species of plant which is, or is likely to be, detrimental or destructive and difficult to control or eradicate” (NRS 555.05). To assist the reader, we also included a separate table detailing the noxious weeds, category level (A, B, or C), and the typical habitats that these species invade. -

WOOD ANATOMY of CHENOPODIACEAE (AMARANTHACEAE S
IAWA Journal, Vol. 33 (2), 2012: 205–232 WOOD ANATOMY OF CHENOPODIACEAE (AMARANTHACEAE s. l.) Heike Heklau1, Peter Gasson2, Fritz Schweingruber3 and Pieter Baas4 SUMMARY The wood anatomy of the Chenopodiaceae is distinctive and fairly uni- form. The secondary xylem is characterised by relatively narrow vessels (<100 µm) with mostly minute pits (<4 µm), and extremely narrow ves- sels (<10 µm intergrading with vascular tracheids in addition to “normal” vessels), short vessel elements (<270 µm), successive cambia, included phloem, thick-walled or very thick-walled fibres, which are short (<470 µm), and abundant calcium oxalate crystals. Rays are mainly observed in the tribes Atripliceae, Beteae, Camphorosmeae, Chenopodieae, Hab- litzieae and Salsoleae, while many Chenopodiaceae are rayless. The Chenopodiaceae differ from the more tropical and subtropical Amaran- thaceae s.str. especially in their shorter libriform fibres and narrower vessels. Contrary to the accepted view that the subfamily Polycnemoideae lacks anomalous thickening, we found irregular successive cambia and included phloem. They are limited to long-lived roots and stem borne roots of perennials (Nitrophila mohavensis) and to a hemicryptophyte (Polycnemum fontanesii). The Chenopodiaceae often grow in extreme habitats, and this is reflected by their wood anatomy. Among the annual species, halophytes have narrower vessels than xeric species of steppes and prairies, and than species of nitrophile ruderal sites. Key words: Chenopodiaceae, Amaranthaceae s.l., included phloem, suc- cessive cambia, anomalous secondary thickening, vessel diameter, vessel element length, ecological adaptations, xerophytes, halophytes. INTRODUCTION The Chenopodiaceae in the order Caryophyllales include annual or perennial herbs, sub- shrubs, shrubs, small trees (Haloxylon ammodendron, Suaeda monoica) and climbers (Hablitzia, Holmbergia). -

An Illustrated Key to the Amaranthaceae of Alberta
AN ILLUSTRATED KEY TO THE AMARANTHACEAE OF ALBERTA Compiled and writen by Lorna Allen & Linda Kershaw April 2019 © Linda J. Kershaw & Lorna Allen This key was compiled using informaton primarily from Moss (1983), Douglas et. al. (1998a [Amaranthaceae], 1998b [Chenopodiaceae]) and the Flora North America Associaton (2008). Taxonomy follows VASCAN (Brouillet, 2015). Please let us know if there are ways in which the key can be improved. The 2015 S-ranks of rare species (S1; S1S2; S2; S2S3; SU, according to ACIMS, 2015) are noted in superscript (S1;S2;SU) afer the species names. For more details go to the ACIMS web site. Similarly, exotc species are followed by a superscript X, XX if noxious and XXX if prohibited noxious (X; XX; XXX) according to the Alberta Weed Control Act (2016). AMARANTHACEAE Amaranth Family [includes Chenopodiaceae] Key to Genera 01a Flowers with spiny, dry, thin and translucent 1a (not green) bracts at the base; tepals dry, thin and translucent; separate ♂ and ♀ fowers on same the plant; annual herbs; fruits thin-walled (utricles), splitting open around the middle 2a (circumscissile) .............Amaranthus 01b Flowers without spiny, dry, thin, translucent bracts; tepals herbaceous or feshy, greenish; fowers various; annual or perennial, herbs or shrubs; fruits various, not splitting open around the middle ..........................02 02a Leaves scale-like, paired (opposite); stems feshy/succulent, with fowers sunk into stem; plants of saline habitats ... Salicornia rubra 3a ................. [Salicornia europaea] 02b Leaves well developed, not scale-like; stems not feshy; plants of various habitats. .03 03a Flower bracts tipped with spine or spine-like bristle; leaves spine-tipped, linear to awl- 5a shaped, usually not feshy; tepals winged from the lower surface .............. -

The Ploidy Races of Atriplex Confertifolia (Chenopodiaceae)
Western North American Naturalist 71(1), © 2011, pp. 67–77 THE PLOIDY RACES OF ATRIPLEX CONFERTIFOLIA (CHENOPODIACEAE) Stewart C. Sanderson1 ABSTRACT.—Previous accounts of polyploidy in the North American salt desert shrub Atriplex confertifolia (shad- scale) have dealt with the distribution of polyploidy and the morphological and secondary chemical differences between races. The present study amplifies these studies and reveals additional ploidy-flavonoid races, with ploidy levels known to extend from 2x to 12x, and all except 2x and 12x represented by races with and without 6-methoxylation of flavonol compounds. Results of this study show that diploids across their range have about 113% as much DNA per genome as do polyploids and that parallel variation in monoploid genome size between diploids and accompanying polyploids can be shown in different parts of the species’ range. Polyploidy, therefore, appears to have developed independently in sev- eral areas of the western United States. Hexaploids are generally not as common as octoploids in shadscale, which could be an indication of diploidization of older tetraploid races. RESUMEN.—Informes anteriores sobre la poliploidía en el arbusto Atriplex confertifolia (“shadscale”) del desierto de sal norteamericano, han tratado la distribución de la poliploidía, las diferencias químicas secundarias y las diferencias morfológi- cas entre razas. El presente estudio amplía los estudios previos y revela razas ploidía-flavonoides adicionales, con niveles de ploidía que se sabe que van de 2x a 12x, y todos, con excepción de 2x y 12x, están representados en razas con y sin 6-metoxi- lación de compuestos de flavonoles. Durante el estudio descubrí que los diploides a lo largo de su rango de distribución tienen alrededor del 113% más ADN por genoma que los polploides. -

DOI: Review Article Atriplex Canescens (Pursh) Nutt
Revista Mexicana de Ciencias Forestales Vol. 12 (67) Septiembre – Octubre (2021) DOI: https://doi.org/10.29298/rmcf.v12i67.821 Review Article Atriplex canescens (Pursh) Nutt. una especie multifuncional de las zonas semiáridas de Norteamérica: una revisión Atriplex canescens (Pursh) Nutt. a multifunctional species of the semi-arid zones of north America: a review David Castillo Quiroz1, Ramón Gutiérrez Luna2, Diana Yemilet Ávila Flores1, Francisco Castillo Reyes1 y Jesús Eduardo Sáenz Ceja3* Resumen Atriplex canescens es una especie nativa ampliamente distribuida en las zonas semiáridas de Norteamérica, desde el norte de México hasta el oeste de Estados Unidos de América. La presente revisión de la información publicada sobre esta especie durante los últimos 25 años pretende mostrar su taxonomía, distribución geográfica, hábitat, usos actuales y potenciales, así como las amenazas para su hábitat. Los resultados evidenciaron que el uso más amplio de A. canescens es el forrajero, en la alimentación de ganado bovino, caprino y ovino. También, destacó su empleo en la rehabilitación de suelos degradados, la captura de carbono, la prevención de la erosión y la fitorremediación de suelos contaminados por desechos industriales. Además, tiene un amplio potencial en el campo biotecnológico, como control biológico, biocombustible y fuente de genes tolerantes a la sequía y salinidad; asimismo, el consumo de flores, frutos, hojas y raíces de A. canescens ha sido muy importante para las comunidades indígenas. Sin embargo, a pesar de su amplia distribución enfrenta algunas amenazas, como el cambio de uso de suelo, la competencia con especies invasoras y la reducción de la conectividad entre poblaciones naturales. En síntesis, A. -

Plants of Havasu National Wildlife Refuge
U.S. Fish and Wildlife Service Plants of Havasu National Wildlife Refuge The Havasu NWR plant list was developed by volunteer Baccharis salicifolia P S N John Hohstadt. As of October 2012, 216 plants have been mulefat documented at the refuge. Baccharis brachyphylla P S N Legend shortleaf baccharis *Occurance (O) *Growth Form (GF) *Exotic (E) Bebbia juncea var. aspera P S N A=Annual G=Grass Y=Yes sweetbush P=Perennial F=Forb N=No Calycoseris wrightii A F N B=Biennial S=Shrub T=Tree white tackstem Calycoseris parryi A F N Family yellow tackstem Scientific Name O* GF* E* Chaenactis carphoclinia A F N Common Name pebble pincushion Agavaceae—Lilies Family Chaenactis fremontii A F N Androstephium breviflorum P F N pincushion flower pink funnel lily Conyza canadensis A F N Hesperocallis undulata P F N Canadian horseweed desert lily Chrysothamnus Spp. P S N Aizoaceae—Fig-marigold Family rabbitbrush Sesuvium sessile A F N Encelia frutescens P S N western seapurslane button brittlebrush Encelia farinosa P S N Aizoaceae—Fig-marigold Family brittlebrush Trianthema portulacastrum A F N Dicoria canescens A F N desert horsepurslane desert twinbugs Amaranthaceae—Amaranth Family Antheropeas wallacei A F N Amaranthus retroflexus A F N woolly easterbonnets redroot amaranth Antheropeas lanosum A F N Tidestromia oblongifolia P F N white easterbonnets Arizona honeysweet Ambrosia dumosa P S N burrobush Apiaceae—Carrot Family Ambrosia eriocentra P S N Bowlesia incana P F N woolly fruit bur ragweed hoary bowlesia Geraea canescens A F N Hydrocotyle verticillata P F N hairy desertsunflower whorled marshpennywort Gnaphalium spp. -

Vascular Plant Species of the Comanche National Grassland in United States Department Southeastern Colorado of Agriculture
Vascular Plant Species of the Comanche National Grassland in United States Department Southeastern Colorado of Agriculture Forest Service Donald L. Hazlett Rocky Mountain Research Station General Technical Report RMRS-GTR-130 June 2004 Hazlett, Donald L. 2004. Vascular plant species of the Comanche National Grassland in southeast- ern Colorado. Gen. Tech. Rep. RMRS-GTR-130. Fort Collins, CO: U.S. Department of Agriculture, Forest Service, Rocky Mountain Research Station. 36 p. Abstract This checklist has 785 species and 801 taxa (for taxa, the varieties and subspecies are included in the count) in 90 plant families. The most common plant families are the grasses (Poaceae) and the sunflower family (Asteraceae). Of this total, 513 taxa are definitely known to occur on the Comanche National Grassland. The remaining 288 taxa occur in nearby areas of southeastern Colorado and may be discovered on the Comanche National Grassland. The Author Dr. Donald L. Hazlett has worked as an ecologist, botanist, ethnobotanist, and teacher in Latin America and in Colorado. He has specialized in the flora of the eastern plains since 1985. His many years in Latin America prompted him to include Spanish common names in this report, names that are seldom reported in floristic pub- lications. He is also compiling plant folklore stories for Great Plains plants. Since Don is a native of Otero county, this project was of special interest. All Photos by the Author Cover: Purgatoire Canyon, Comanche National Grassland You may order additional copies of this publication by sending your mailing information in label form through one of the following media. -

Desert Plants of Utah
DESERT PLANTS OF UTAH Original booklet and drawings by Berniece A. Andersen Revised May 1996 HG 505 FOREWORD The original Desert Plants of Utah by Berniece A. Andersen has been a remarkably popular book, serving as a tribute to both her botanical knowledge of the region and to her enthusiastic manner. For these reasons, we have tried to retain as much of the original work, in both content and style, as possible. Some modifications were necessary. We have updated scientific names in accordance with changes that have occurred since the time of the first publication and we have also incorporated new geographic distributional data that have accrued with additional years of botanical exploration. The most obvious difference pertains to the organization of species. In the original version, species were organized phylogenetically, reflecting the predominant concepts of evolutionary relationships among plant families at that time. In an effort to make this version more user-friendly for the beginner, we have chosen to arrange the plants primarily by flower color. We hope that these changes will not diminish the enjoyment gained by anyone familiar with the original. We would also like to thank Larry A. Rupp, Extension Horticulture Specialist, for critical review of the draft and for the cover photo. Linda Allen, Assistant Curator, Intermountain Herbarium Donna H. Falkenborg, Extension Editor TABLE OF CONTENTS The Nature of Deserts ........................................................1 Utah’s Deserts ........................................................2 -

Checklist of the Vascular Plants of San Diego County 5Th Edition
cHeckliSt of tHe vaScUlaR PlaNtS of SaN DieGo coUNty 5th edition Pinus torreyana subsp. torreyana Downingia concolor var. brevior Thermopsis californica var. semota Pogogyne abramsii Hulsea californica Cylindropuntia fosbergii Dudleya brevifolia Chorizanthe orcuttiana Astragalus deanei by Jon P. Rebman and Michael G. Simpson San Diego Natural History Museum and San Diego State University examples of checklist taxa: SPecieS SPecieS iNfRaSPecieS iNfRaSPecieS NaMe aUtHoR RaNk & NaMe aUtHoR Eriodictyon trichocalyx A. Heller var. lanatum (Brand) Jepson {SD 135251} [E. t. subsp. l. (Brand) Munz] Hairy yerba Santa SyNoNyM SyMBol foR NoN-NATIVE, NATURaliZeD PlaNt *Erodium cicutarium (L.) Aiton {SD 122398} red-Stem Filaree/StorkSbill HeRBaRiUM SPeciMeN coMMoN DocUMeNTATION NaMe SyMBol foR PlaNt Not liSteD iN THE JEPSON MANUAL †Rhus aromatica Aiton var. simplicifolia (Greene) Conquist {SD 118139} Single-leaF SkunkbruSH SyMBol foR StRict eNDeMic TO SaN DieGo coUNty §§Dudleya brevifolia (Moran) Moran {SD 130030} SHort-leaF dudleya [D. blochmaniae (Eastw.) Moran subsp. brevifolia Moran] 1B.1 S1.1 G2t1 ce SyMBol foR NeaR eNDeMic TO SaN DieGo coUNty §Nolina interrata Gentry {SD 79876} deHeSa nolina 1B.1 S2 G2 ce eNviRoNMeNTAL liStiNG SyMBol foR MiSiDeNtifieD PlaNt, Not occURRiNG iN coUNty (Note: this symbol used in appendix 1 only.) ?Cirsium brevistylum Cronq. indian tHiStle i checklist of the vascular plants of san Diego county 5th edition by Jon p. rebman and Michael g. simpson san Diego natural history Museum and san Diego state university publication of: san Diego natural history Museum san Diego, california ii Copyright © 2014 by Jon P. Rebman and Michael G. Simpson Fifth edition 2014. isBn 0-918969-08-5 Copyright © 2006 by Jon P.