The Cardioprotective Effects of the New Crystal Form of Puerarin in Isoproterenol-Induced Myocardial Ischemia Rats Based on Meta
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
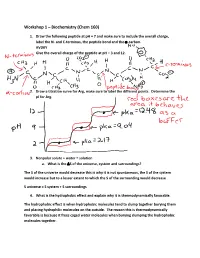
Workshop 1 – Biochemistry (Chem 160)
Workshop 1 – Biochemistry (Chem 160) 1. Draw the following peptide at pH = 7 and make sure to include the overall charge, label the N- and C-terminus, the peptide bond and the -carbon. AVDKY Give the overall charge of the peptide at pH = 3 and 12. 2. Draw a titration curve for Arg, make sure to label the different points. Determine the pI for Arg. 3. Nonpolar solute + water = solution a. What is the S of the universe, system and surroundings? The S of the universe would decrease this is why it is not spontaneous, the S of the system would increase but to a lesser extent to which the S of the surrounding would decrease S universe = S system + S surroundings 4. What is the hydrophobic effect and explain why it is thermodynamically favorable. The hydrophobic effect is when hydrophobic molecules tend to clump together burying them and placing hydrophilic molecules on the outside. The reason this is thermodynamically favorable is because it frees caged water molecules when burying clumping the hydrophobic molecules together. 5. Urea dissolves very readily in water, but the solution becomes very cold as the urea dissolves. How is this possible? Urea dissolves in water because when dissolving there is a net increase in entropy of the universe. The heat exchange, getting colder only reflects the enthalpy (H) component of the total energy change. The entropy change is high enough to offset the enthalpy component and to add up to an overall -G 6. A mutation that changes an alanine residue in the interior of a protein to valine is found to lead to a loss of activity. -

Amino Acid Recognition by Aminoacyl-Trna Synthetases
www.nature.com/scientificreports OPEN The structural basis of the genetic code: amino acid recognition by aminoacyl‑tRNA synthetases Florian Kaiser1,2,4*, Sarah Krautwurst3,4, Sebastian Salentin1, V. Joachim Haupt1,2, Christoph Leberecht3, Sebastian Bittrich3, Dirk Labudde3 & Michael Schroeder1 Storage and directed transfer of information is the key requirement for the development of life. Yet any information stored on our genes is useless without its correct interpretation. The genetic code defnes the rule set to decode this information. Aminoacyl-tRNA synthetases are at the heart of this process. We extensively characterize how these enzymes distinguish all natural amino acids based on the computational analysis of crystallographic structure data. The results of this meta-analysis show that the correct read-out of genetic information is a delicate interplay between the composition of the binding site, non-covalent interactions, error correction mechanisms, and steric efects. One of the most profound open questions in biology is how the genetic code was established. While proteins are encoded by nucleic acid blueprints, decoding this information in turn requires proteins. Te emergence of this self-referencing system poses a chicken-or-egg dilemma and its origin is still heavily debated 1,2. Aminoacyl-tRNA synthetases (aaRSs) implement the correct assignment of amino acids to their codons and are thus inherently connected to the emergence of genetic coding. Tese enzymes link tRNA molecules with their amino acid cargo and are consequently vital for protein biosynthesis. Beside the correct recognition of tRNA features3, highly specifc non-covalent interactions in the binding sites of aaRSs are required to correctly detect the designated amino acid4–7 and to prevent errors in biosynthesis5,8. -

Amino Acid Chemistry
Handout 4 Amino Acid and Protein Chemistry ANSC 619 PHYSIOLOGICAL CHEMISTRY OF LIVESTOCK SPECIES Amino Acid Chemistry I. Chemistry of amino acids A. General amino acid structure + HN3- 1. All amino acids are carboxylic acids, i.e., they have a –COOH group at the #1 carbon. 2. All amino acids contain an amino group at the #2 carbon (may amino acids have a second amino group). 3. All amino acids are zwitterions – they contain both positive and negative charges at physiological pH. II. Essential and nonessential amino acids A. Nonessential amino acids: can make the carbon skeleton 1. From glycolysis. 2. From the TCA cycle. B. Nonessential if it can be made from an essential amino acid. 1. Amino acid "sparing". 2. May still be essential under some conditions. C. Essential amino acids 1. Branched chain amino acids (isoleucine, leucine and valine) 2. Lysine 3. Methionine 4. Phenyalanine 5. Threonine 6. Tryptophan 1 Handout 4 Amino Acid and Protein Chemistry D. Essential during rapid growth or for optimal health 1. Arginine 2. Histidine E. Nonessential amino acids 1. Alanine (from pyruvate) 2. Aspartate, asparagine (from oxaloacetate) 3. Cysteine (from serine and methionine) 4. Glutamate, glutamine (from α-ketoglutarate) 5. Glycine (from serine) 6. Proline (from glutamate) 7. Serine (from 3-phosphoglycerate) 8. Tyrosine (from phenylalanine) E. Nonessential and not required for protein synthesis 1. Hydroxyproline (made postranslationally from proline) 2. Hydroxylysine (made postranslationally from lysine) III. Acidic, basic, polar, and hydrophobic amino acids A. Acidic amino acids: amino acids that can donate a hydrogen ion (proton) and thereby decrease pH in an aqueous solution 1. -

Solutions to 7.012 Problem Set 1
MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Solutions to 7.012 Problem Set 1 Question 1 Bob, a student taking 7.012, looks at a long-standing puddle outside his dorm window. Curious as to what was growing in the cloudy water, he takes a sample to his TA, Brad Student. He wanted to know whether the organisms in the sample were prokaryotic or eukaryotic. a) Give an example of a prokaryotic and a eukaryotic organism. Prokaryotic: Eukaryotic: All bacteria Yeast, fungi, any animial or plant b) Using a light microscope, how could he tell the difference between a prokaryotic organism and a eukaryotic one? The resolution of the light microscope would allow you to see if the cell had a true nucleus or organelles. A cell with a true nucleus and organelles would be eukaryotic. You could also determine size, but that may not be sufficient to establish whether a cell is prokaryotic or eukaryotic. c) What additional differences exist between prokaryotic and eukaryotic organisms? Any answer from above also fine here. In addition, prokaryotic and eukaryotic organisms differ at the DNA level. Eukaryotes have more complex genomes than prokaryotes do. Question 2 A new startup company hires you to help with their product development. Your task is to find a protein that interacts with a polysaccharide. a) You find a large protein that has a single binding site for the polysaccharide cellulose. Which amino acids might you expect to find in the binding pocket of the protein? What is the strongest type of interaction possible between these amino acids and the cellulose? Cellulose is a polymer of glucose and as such has many free hydroxyl groups. -

Nucleotide Base Coding and Am1ino Acid Replacemients in Proteins* by Emil L
VOL. 48, 1962 BIOCHEMISTRY: E. L. SAIITH 677 18 Britten, R. J., and R. B. Roberts, Science, 131, 32 (1960). '9 Crestfield, A. M., K. C. Smith, and F. WV. Allen, J. Biol. Chem., 216, 185 (1955). 20 Gamow, G., Nature, 173, 318 (1954). 21 Brenner, S., these PROCEEDINGS, 43, 687 (1957). 22 Nirenberg, M. WV., J. H. Matthaei, and 0. WV. Jones, unpublished data. 23 Crick, F. H. C., L. Barnett, S. Brenner, and R. J. Watts-Tobin, Nature, 192, 1227 (1961). 24 Levene, P. A., and R. S. Tipson, J. Biol. Ch-nn., 111, 313 (1935). 25 Gierer, A., and K. W. Mundry, Nature, 182, 1437 (1958). 2' Tsugita, A., and H. Fraenkel-Conrat, J. Mllot. Biol., in press. 27 Tsugita, A., and H. Fraenkel-Conrat, personal communication. 28 Wittmann, H. G., Naturwissenschaften, 48, 729 (1961). 29 Freese, E., in Structure and Function of Genetic Elements, Brookhaven Symposia in Biology, no. 12 (1959), p. 63. NUCLEOTIDE BASE CODING AND AM1INO ACID REPLACEMIENTS IN PROTEINS* BY EMIL L. SMITHt LABORATORY FOR STUDY OF HEREDITARY AND METABOLIC DISORDERS AND THE DEPARTMENTS OF BIOLOGICAL CHEMISTRY AND MEDICINE, UNIVERSITY OF UTAH COLLEGE OF MEDICINE Communicated by Severo Ochoa, February 14, 1962 The problem of which bases of messenger or template RNA' specify the coding of amino acids in proteins has been largely elucidated by the use of synthetic polyri- bonucleotides.2-7 For these triplet nucleotide compositions (Table 1), it is of in- terest to examine some of the presently known cases of amino acid substitutions in polypeptides or proteins of known structure. -

Amino Acid Requirements of the Free-Living Nematode Caenorhabditis Briggsae
AMINO ACID REQUIREMENTS OF THE FREE-LIVING NEMATODE CAENORHABDITIS BRIGGSAE BY J. R. VANFLETEREN Instituut voor Dierkunde, Laboratoria voor Morfologie en Systematiek, RijksuniversiteitGent, Belgium Washed yeast ribosomes promote growth and reproduction of C. briggsae, even when supple- mented to the basal medium at dosages too low to provide the organisms with sufficient amounts of essential amino acids. Hence, a re-investigation of the amino acid requirements of C. briggsae by single and multiple omission of amino acids from the basal medium revealed unambiguously that arginine, histidine, lysine, tryptophan, phenylalanine, methionine, threonine, leucine, isoleucine and valine are not synthetized at levels to permit reproduction; they are called essential amino acids. The requirement for arginine and isoleucinehowever appears to be less clear-cut. On the contrary, evidence is presented that alanine, asparagine, cysteine, glutamate, glutamine, glycine, proline, serine and tryosine can be synthetized at adequate levels; they are called non- essential amino acids. In addition it was shown that multiple omission of the non-essential amino acids is not deleterious. This is believed to be an important step towards the development of a minimum essential medium (MEM) for growth and reproduction of C. briggsae. Sustained growth of the free-living nematode Caenorhabditis brigg.rae can be obtained on a chemically defined medium, supplemented with adequate levels of a proteinaceous growth factor. The most satisfactory, chemically defined medium hitherto reported (Buecher, Hansen & Yarwood, 1966), has been called C. brigg.iae Maintenance Medium (CbMM) and is now commercially available. CbMM is an extremely rich medium, being composed of 53 components, all present at high concentrations. -
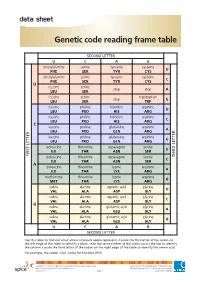
Genetic Code Reading Frame Table
data sheet Genetic code reading frame table SECOND LETTER U C A G phenylalanine serine tyrosine cysteine U PHE SER TYR CYS phenylalanine serine tyrosine cysteine C PHE SER TYR CYS U leucine serine stop stop A LEU SER leucine serine tryptophan stop G LEU SER TRP leucine proline histidine arginine U LEU PRO HIS ARG leucine proline histidine arginine C LEU PRO HIS ARG C leucine proline glutamine arginine A LEU PRO GLN ARG leucine proline glutamine arginine G LEU PRO GLN ARG isoleucine threonine asparagine serine U ILE THR ASN SER FIRST LETTER isoleucine threonine asparagine serine LETTERTHIRD C ILE THR ASN SER A isoleucine threonine lysine arginine A ILE THR LYS ARG methionine threonine lysine arginine G MET THR LYS ARG valine alanine aspartic acid glycine U VAL ALA ASP GLY valine alanine aspartic acid glycine C VAL ALA ASP GLY G valine alanine glutamic acid glycine A VAL ALA GLU GLY valine alanine glutamic acid glycine G VAL ALA GLU GLY U C A G SECOND LETTER Use this table to find out what amino acid each codon represents. Locate the first letter of the codon on the left edge of the table to identify a block, then the second letter of the codon across the top to identify the column. Locate the third letter of the codon on the right edge of the table to identify the amino acid. For example, the codon ‘CAU’ codes for histidine (HIS). ast0812 | Proteins 3: Genetic code reading frame table (data sheet) developed for the Department of Education WA © The University of Western Australia 2012 for conditions of use see spice.wa.edu.au/usage version 1.0 page 1 Licensed for NEALS. -

Amino Acid Degradation
BI/CH 422/622 OUTLINE: OUTLINE: Protein Degradation (Catabolism) Digestion Amino-Acid Degradation Inside of cells Protein turnover Dealing with the carbon Ubiquitin Fates of the 29 Activation-E1 Seven Families Conjugation-E2 nitrogen atoms in 20 1. ADENQ Ligation-E3 AA: Proteosome 2. RPH 9 ammonia oxidase Amino-Acid Degradation 18 transamination Ammonia 2 urea one-carbon metabolism free transamination-mechanism to know THF Urea Cycle – dealing with the nitrogen SAM 5 Steps Carbamoyl-phosphate synthetase 3. GSC Ornithine transcarbamylase PLP uses Arginino-succinate synthetase Arginino-succinase 4. MT – one carbon metabolism Arginase 5. FY – oxidase vs oxygenase Energetics Urea Bi-cycle 6. KW – Urea Cycle – dealing with the nitrogen 7. BCAA – VIL Feeding the Urea Cycle Glucose-Alanine Cycle Convergence with Fatty acid-odd chain Free Ammonia Overview Glutamine Glutamate dehydrogenase Overall energetics Amino Acid A. Concepts 1. ConvergentDegradation 2. ketogenic/glucogenic 3. Reactions seen before The SEVEN (7) Families B. Transaminase (A,D,E) / Deaminase (Q,N) Family C. Related to biosynthesis (R,P,H; C,G,S; M,T) 1.Glu Family a. Introduce oxidases/oxygenases b. Introduce one-carbon metabolism (1C) 2.Pyruvate Family a. PLP reactions 3. a-Ketobutyric Family (M,T) a. 1-C metabolism D. Dedicated 1. Aromatic Family (F,Y) a. oxidases/oxygenases 2. a-Ketoadipic Family (K,W) 3. Branched-chain Family (V,I,L) E. Convergence with Fatty Acids: propionyl-CoA 29 N 1 Amino Acid Degradation • Intermediates of the central metabolic pathway • Some amino acids result in more than one intermediate. • Ketogenic amino acids can be converted to ketone bodies. -

Isolation, Identification, and Studies on the Metabolism of Rumen Micro-Organism Growth Factors Present in Natural Materials
ISOLATION, IDENTIFICATION, AND STUDIES ON THE METABOLISM OF RUMEN MICRO-ORGANISM GROWTH FACTORS PRESENT IN NATURAL MATERIALS DISSERTATION Presented In Partial Fuirillraent of the Requirements for the Degree Doctor of Philosophy in the Graduate School of the Ohio State University By BURK ALLYN DEHOR ITT, A.B., M.S. The Ohio State University 1957 Approved by: Adviser Department of Agricultural Biochemistry ACKNOWLEDGMENTS I would like to express ray deepest appreciation to Dr. Alvin L. Moxon and Dr. Orville G. Bentley for their guidance and many helpful suggestions during the per formance of ray research work. Their interest and leader ship is gratefully acknowledged. The timely suggestions and interest of Dr. Ronald R. Johnson are greatly appreciated, in addition to his aid in the preparation of Figures 10, 11, and 12. Appreciation is also extended to the Ohio Agricultural Experiment Station for providing facilities and financial assistance during the course of this work. I am indebted to ray wife for her unceasing encour agement and cheerfulness throughout the performance of this work, and her aid in the preparation of this manu script. ii TABLE OP CONTENTS General Introduction Part I: Isolation and .identification of Compounds Prom Autolyzed Yeast, Alfalfa Ileal, and Casoin iiydrolysa.te wit h. Cellulolytic Factor Activity for Rumen Micro-organisms In Vitro Literature Review Experimental Procedures In Vitro Rumen ^Fermentation Technique Fractionation Procedures Dowex - 50 I o n Exchange Resin Charcoal Treatment Preparative Scale -

24Amino Acids, Peptides, and Proteins
WADEMC24_1153-1199hr.qxp 16-12-2008 14:15 Page 1153 CHAPTER COOϪ a -h eli AMINO ACIDS, x ϩ PEPTIDES, AND NH3 PROTEINS Proteins are the most abundant organic molecules 24-1 in animals, playing important roles in all aspects of cell structure and function. Proteins are biopolymers of Introduction 24A-amino acids, so named because the amino group is bonded to the a carbon atom, next to the carbonyl group. The physical and chemical properties of a protein are determined by its constituent amino acids. The individual amino acid subunits are joined by amide linkages called peptide bonds. Figure 24-1 shows the general structure of an a-amino acid and a protein. α carbon atom O H2N CH C OH α-amino group R side chain an α-amino acid O O O O O H2N CH C OH H2N CH C OH H2N CH C OH H2N CH C OH H2N CH C OH CH3 CH2OH H CH2SH CH(CH3)2 alanine serine glycine cysteine valine several individual amino acids peptide bonds O O O O O NH CH C NH CH C NH CH C NH CH C NH CH C CH3 CH2OH H CH2SH CH(CH3)2 a short section of a protein a FIGURE 24-1 Structure of a general protein and its constituent amino acids. The amino acids are joined by amide linkages called peptide bonds. 1153 WADEMC24_1153-1199hr.qxp 16-12-2008 14:15 Page 1154 1154 CHAPTER 24 Amino Acids, Peptides, and Proteins TABLE 24-1 Examples of Protein Functions Class of Protein Example Function of Example structural proteins collagen, keratin strengthen tendons, skin, hair, nails enzymes DNA polymerase replicates and repairs DNA transport proteins hemoglobin transports O2 to the cells contractile proteins actin, myosin cause contraction of muscles protective proteins antibodies complex with foreign proteins hormones insulin regulates glucose metabolism toxins snake venoms incapacitate prey Proteins have an amazing range of structural and catalytic properties as a result of their varying amino acid composition. -

Metabolism of Valine and the Exchange of Amino Acids Across the Hind-Limb Muscles of Fed and Starved Sheep
Aust. 1. BioI. Sci., 1986, 39, 379-93 Metabolism of Valine and the Exchange of Amino Acids across the Hind-limb Muscles of Fed and Starved Sheep E. Teleni, A,B E. F. Annison A and D. B. LindsayA,C A Department of Animal Husbandry, University of Sydney, Camden, N.S.W. 2570. B Present address: Graduate School of Tropical Veterinary Science, James Cook University, Townsville, Qld 4811. C Present address: Tropical Cattle Centre, CSIRO, Bruce Highway, North Rockhampton, Qld 4702. Abstract A combination of the isotope-dilution and arterio-venous (A V) difference techniques was used to study simultaneously the metabolism of valine in the whole body and in the hind-limb muscles of fed and starved (40 h) sheep. The net exchange of gluconeogenic amino acids across hind-limb muscles was also studied. Valine entry rate was unaffected by nutritional status. There was significant extraction of valine by hind-limb muscles in both fed and starved sheep. The percentage of valine uptake decarboxylated was higher (P < 0'05) in fed sheep but the amount of valine decarboxylated was not significantly different. The proportion of valine uptake that was transaminated was about 30 times higher in starved sheep. About 54% of valine taken up by hind-limb muscle of starved sheep was metabolized. The corresponding value for fed sheep was 21 %. The contribution of CO2 from valine decarboxylation to total hind-limb muscle C02 output was about 0·2%. The output of alanine in both fed and starved sheep was low but the output of glutamine was relatively high and roughly equivalent to the amounts of aspartate, glutamate and branched-chain amino acids that were catabolized. -

Combined Effect of Arginine, Valine, and Serine on Exercise
nutrients Article Combined Effect of Arginine, Valine, and Serine on Exercise-Induced Fatigue in Healthy Volunteers: A Randomized, Double-Blinded, Placebo-Controlled Crossover Study Yuichi Tsuda *, Makoto Yamaguchi, Teruyuki Noma, Eiji Okaya and Hiroyuki Itoh R&D Division, Meiji Co., Ltd., 1-29-1 Nanakuni, Hachiouji, Tokyo 192-0919, Japan; [email protected] (M.Y.); [email protected] (T.N.); [email protected] (E.O.); [email protected] (H.I.) * Correspondence: [email protected]; Tel.: +81-42-632-5849 Received: 2 April 2019; Accepted: 15 April 2019; Published: 17 April 2019 Abstract: Although several kinds of amino acids (AAs) are known to affect physiological actions during exercise, little is known about the combined effects of a mixture of several AAs on fatigue during exercise. The aim of the present study was to investigate the effect of an AA mixture supplement containing arginine, valine, and serine on exercise-induced fatigue in healthy volunteers. These AAs were selected because they were expected to reduce fatigue during exercise by acting the positive effects synergistically. A randomized, double-blinded, placebo-controlled crossover trial was conducted. Thirty-nine males ingested an AA mixture containing 3600 mg of arginine, 2200 mg of valine, and 200 mg of serine or a placebo each day for 14 days. On the 14th day, the participants completed an exercise trial on a cycle ergometer at 50% of VO2max for 120 min. After the two-week washout period, the participants repeated the same trial with the other test sample. The participant’s feeling of fatigue based on a visual analog scale (VAS) and a rating of perceived exertion (RPE), as well as blood and physical parameters were evaluated.