Nucleotide Base Coding and Am1ino Acid Replacemients in Proteins* by Emil L
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
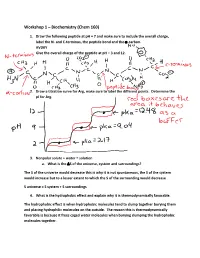
Workshop 1 – Biochemistry (Chem 160)
Workshop 1 – Biochemistry (Chem 160) 1. Draw the following peptide at pH = 7 and make sure to include the overall charge, label the N- and C-terminus, the peptide bond and the -carbon. AVDKY Give the overall charge of the peptide at pH = 3 and 12. 2. Draw a titration curve for Arg, make sure to label the different points. Determine the pI for Arg. 3. Nonpolar solute + water = solution a. What is the S of the universe, system and surroundings? The S of the universe would decrease this is why it is not spontaneous, the S of the system would increase but to a lesser extent to which the S of the surrounding would decrease S universe = S system + S surroundings 4. What is the hydrophobic effect and explain why it is thermodynamically favorable. The hydrophobic effect is when hydrophobic molecules tend to clump together burying them and placing hydrophilic molecules on the outside. The reason this is thermodynamically favorable is because it frees caged water molecules when burying clumping the hydrophobic molecules together. 5. Urea dissolves very readily in water, but the solution becomes very cold as the urea dissolves. How is this possible? Urea dissolves in water because when dissolving there is a net increase in entropy of the universe. The heat exchange, getting colder only reflects the enthalpy (H) component of the total energy change. The entropy change is high enough to offset the enthalpy component and to add up to an overall -G 6. A mutation that changes an alanine residue in the interior of a protein to valine is found to lead to a loss of activity. -

Effects of Single Amino Acid Deficiency on Mrna Translation Are Markedly
www.nature.com/scientificreports OPEN Efects of single amino acid defciency on mRNA translation are markedly diferent for methionine Received: 12 December 2016 Accepted: 4 May 2018 versus leucine Published: xx xx xxxx Kevin M. Mazor, Leiming Dong, Yuanhui Mao, Robert V. Swanda, Shu-Bing Qian & Martha H. Stipanuk Although amino acids are known regulators of translation, the unique contributions of specifc amino acids are not well understood. We compared efects of culturing HEK293T cells in medium lacking either leucine, methionine, histidine, or arginine on eIF2 and 4EBP1 phosphorylation and measures of mRNA translation. Methionine starvation caused the most drastic decrease in translation as assessed by polysome formation, ribosome profling, and a measure of protein synthesis (puromycin-labeled polypeptides) but had no signifcant efect on eIF2 phosphorylation, 4EBP1 hyperphosphorylation or 4EBP1 binding to eIF4E. Leucine starvation suppressed polysome formation and was the only tested condition that caused a signifcant decrease in 4EBP1 phosphorylation or increase in 4EBP1 binding to eIF4E, but efects of leucine starvation were not replicated by overexpressing nonphosphorylatable 4EBP1. This suggests the binding of 4EBP1 to eIF4E may not by itself explain the suppression of mRNA translation under conditions of leucine starvation. Ribosome profling suggested that leucine deprivation may primarily inhibit ribosome loading, whereas methionine deprivation may primarily impair start site recognition. These data underscore our lack of a full -

Untargeted Metabolomics Uncovers the Essential Lysine Transporter in Toxoplasma Gondii
H OH metabolites OH Article Untargeted Metabolomics Uncovers the Essential Lysine Transporter in Toxoplasma gondii Joachim Kloehn 1,*,† , Matteo Lunghi 1,†, Emmanuel Varesio 2 , David Dubois 1 and Dominique Soldati-Favre 1,* 1 Department of Microbiology and Molecular Medicine, University of Geneva, CMU, Rue Michel-Servet 1, 1211 Geneva, Switzerland; [email protected] (M.L.); [email protected] (D.D.) 2 Institute of Pharmaceutical Sciences of Western Switzerland, School of Pharmaceutical Sciences, Mass Spectrometry Core Facility (MZ 2.0), University of Geneva, 1211 Geneva, Switzerland; [email protected] * Correspondence: [email protected] (J.K.); [email protected] (D.S.-F.); Tel.: +41-22-379-57-16 (J.K.); +41-22-379-56-72 (D.S.-F.) † These authors contributed equally to the work. Abstract: Apicomplexan parasites are responsible for devastating diseases, including malaria, toxo- plasmosis, and cryptosporidiosis. Current treatments are limited by emerging resistance to, as well as the high cost and toxicity of existing drugs. As obligate intracellular parasites, apicomplexans rely on the uptake of many essential metabolites from their host. Toxoplasma gondii, the causative agent of tox- oplasmosis, is auxotrophic for several metabolites, including sugars (e.g., myo-inositol), amino acids (e.g., tyrosine), lipidic compounds and lipid precursors (cholesterol, choline), vitamins, cofactors (thiamine) and others. To date, only few apicomplexan metabolite transporters have been charac- terized and assigned a substrate. Here, we set out to investigate whether untargeted metabolomics can be used to identify the substrate of an uncharacterized transporter. Based on existing genome- Citation: Kloehn, J.; Lunghi, M.; and proteome-wide datasets, we have identified an essential plasma membrane transporter of the Varesio, E.; Dubois, D.; Soldati-Favre, major facilitator superfamily in T. -

Amino Acid Recognition by Aminoacyl-Trna Synthetases
www.nature.com/scientificreports OPEN The structural basis of the genetic code: amino acid recognition by aminoacyl‑tRNA synthetases Florian Kaiser1,2,4*, Sarah Krautwurst3,4, Sebastian Salentin1, V. Joachim Haupt1,2, Christoph Leberecht3, Sebastian Bittrich3, Dirk Labudde3 & Michael Schroeder1 Storage and directed transfer of information is the key requirement for the development of life. Yet any information stored on our genes is useless without its correct interpretation. The genetic code defnes the rule set to decode this information. Aminoacyl-tRNA synthetases are at the heart of this process. We extensively characterize how these enzymes distinguish all natural amino acids based on the computational analysis of crystallographic structure data. The results of this meta-analysis show that the correct read-out of genetic information is a delicate interplay between the composition of the binding site, non-covalent interactions, error correction mechanisms, and steric efects. One of the most profound open questions in biology is how the genetic code was established. While proteins are encoded by nucleic acid blueprints, decoding this information in turn requires proteins. Te emergence of this self-referencing system poses a chicken-or-egg dilemma and its origin is still heavily debated 1,2. Aminoacyl-tRNA synthetases (aaRSs) implement the correct assignment of amino acids to their codons and are thus inherently connected to the emergence of genetic coding. Tese enzymes link tRNA molecules with their amino acid cargo and are consequently vital for protein biosynthesis. Beside the correct recognition of tRNA features3, highly specifc non-covalent interactions in the binding sites of aaRSs are required to correctly detect the designated amino acid4–7 and to prevent errors in biosynthesis5,8. -

Hydroxy–Methyl Butyrate (HMB) As an Epigenetic Regulator in Muscle
H OH metabolites OH Communication The Leucine Catabolite and Dietary Supplement β-Hydroxy-β-Methyl Butyrate (HMB) as an Epigenetic Regulator in Muscle Progenitor Cells Virve Cavallucci 1,2,* and Giovambattista Pani 1,2,* 1 Fondazione Policlinico Universitario A. Gemelli IRCCS, 00168 Roma, Italy 2 Institute of General Pathology, Università Cattolica del Sacro Cuore, 00168 Roma, Italy * Correspondence: [email protected] (V.C.); [email protected] (G.P.) Abstract: β-Hydroxy-β-Methyl Butyrate (HMB) is a natural catabolite of leucine deemed to play a role in amino acid signaling and the maintenance of lean muscle mass. Accordingly, HMB is used as a dietary supplement by sportsmen and has shown some clinical effectiveness in preventing muscle wasting in cancer and chronic lung disease, as well as in age-dependent sarcopenia. However, the molecular cascades underlying these beneficial effects are largely unknown. HMB bears a significant structural similarity with Butyrate and β-Hydroxybutyrate (βHB), two compounds recognized for important epigenetic and histone-marking activities in multiple cell types including muscle cells. We asked whether similar chromatin-modifying actions could be assigned to HMB as well. Exposure of murine C2C12 myoblasts to millimolar concentrations of HMB led to an increase in global histone acetylation, as monitored by anti-acetylated lysine immunoblotting, while preventing myotube differentiation. In these effects, HMB resembled, although with less potency, the histone Citation: Cavallucci, V.; Pani, G. deacetylase (HDAC) inhibitor Sodium Butyrate. However, initial studies did not confirm a direct The Leucine Catabolite and Dietary inhibitory effect of HMB on HDACs in vitro. β-Hydroxybutyrate, a ketone body produced by the Supplement β-Hydroxy-β-Methyl liver during starvation or intense exercise, has a modest effect on histone acetylation of C2C12 Butyrate (HMB) as an Epigenetic Regulator in Muscle Progenitor Cells. -

Asparagine-Proline Sequence Within Membrane-Spanning Segment of SREBP Triggers Intramembrane Cleavage by Site-2 Protease
Asparagine-proline sequence within membrane-spanning segment of SREBP triggers intramembrane cleavage by Site-2 protease Jin Ye*†, Utpal P. Dave´ *†, Nick V. Grishin‡, Joseph L. Goldstein*§, and Michael S. Brown*§ Departments of *Molecular Genetics and ‡Biochemistry, University of Texas Southwestern Medical Center, 5323 Harry Hines Boulevard, Dallas, TX 75390-9046 Contributed by Joseph L. Goldstein, March 16, 2000 The NH2-terminal domains of membrane-bound sterol regulatory nus. It translocates to the nucleus, where it activates more than element-binding proteins (SREBPs) are released into the cytosol by 20 genes encoding enzymes of cholesterol and fatty acid synthesis regulated intramembrane proteolysis, after which they enter the as well as the low density lipoprotein receptor (6, 7). When nucleus to activate genes encoding lipid biosynthetic enzymes. sterols build up in cells, the SREBP͞SCAP complex fails to exit Intramembrane proteolysis is catalyzed by Site-2 protease (S2P), a the ER, and it never reaches S1P (8, 9). As a result, the hydrophobic zinc metalloprotease that cleaves SREBPs at a mem- NH2-terminal domains of the SREBPs are no longer released brane-embedded leucine-cysteine bond. In the current study, we into the nucleus, and transcription of the target genes declines. use domain-swapping methods to localize the residues within This mechanism allows cholesterol to inhibit its own synthesis the SREBP-2 membrane-spanning segment that are required for and uptake, thereby preventing cholesterol overaccumulation in cleavage by S2P. The studies reveal a requirement for an asparag- cells. ine-proline sequence in the middle third of the transmembrane The human gene encoding S2P was cloned by complementa- segment. -

Effect of Leucine on Intestinal Absorption of Tryptophan in Rats
Downloaded from https://doi.org/10.1079/BJN19850155 British Journal of Nutrition (1985), 54, 695-703 695 https://www.cambridge.org/core Effect of leucine on intestinal absorption of tryptophan in rats BY CHISAE UMEZAWA, YUKO MAEDA, KANJI HABA, MARIKO SHIN AND KEIJI SANO School of Pharmacy, Kobe-Gakuin University, Nishi-ku, Kobe 673, Japan (Received I7 May 1985 - Accepted 24 June 1985) . IP address: 1. To elucidate the causal relation between leucine and the lowering of hepatic NAD content of rats fed on a leucine-excessive diet (Yamada et aZ. 1979), the effect of leucine on intestinal absorption of tryptophan was 170.106.35.93 investigated. 2. Co-administration of [3H]tryptophan and leucine, with leucine at ten times the level of tryptophan, delayed absorption of L-[side chain 2,3-3H]tryptophan from the digestive tract and incorporation of [3H]tryptophan into portal blood, the liver and a protein fraction of the liver. After 120 min, more than 95% of tryptophan was absorbed whether [3H]tryptophan was administered with or without leucine. , on 3. Co-administration of a mixture of ten essential amino acids, in proportions simulating casein, with 02 Oct 2021 at 04:49:27 [3H]tryptophan markedly delayed absorption of tryptophan from the digestive tract. The addition of supplementary leucine to the amino acid mixture, however, caused no further delay. 4. In rats prefed a leucine-excessive diet for 1 week [3H]tryptophan was absorbed at the same rate as in rats fed on a control diet. 5. The results indicate that competition between tryptophan and leucine for intestinal absorption did not cause lowering of hepatic NAD. -

Electrochemical Studies of Dl-Leucine, L-Proline and L
ELECTROCHEMICAL STUDIES OF DL -LEUCINE, 60 L-PROLINE AND L-TRYPTOPHAN AND THEIR INTERACTION WITH COPPER AND IRON 30 c b A) M. A. Jabbar, R. J. Mannan, S. Salauddin and B. µ a Rashid 0 Department of Chemistry, University of Dhaka, ( Current Dhaka-1000, Bangladesh -30 Introduction -60 In vitro study of the charge transfer reactions coupled -800 -400 0 400 800 with chemical reactions can give important indication of Potential vs. Ag/AgCl (mV) about actual biological processes occurring in human Fig.1. Comparison of the cyclic voltammogram of system. Understanding of such charge-transfer 5.0mM (a) DL -Leucine, (b) Cu-DL -Leucine ion and mechanism will help to determine the effectiveness of (c) [Fe-DL -Leucine] in 0.1M KCl solution at a Pt- nutrition, metabolism and treatment of various biological button electrode. Scan rates 50 mV/s. disorders. In the previous research, the redox behaviour of 40 various amino acids and biochemically important compounds and their charge transfer reaction and their b interaction of metal ions were studied [1,2]. In the present 20 ) a c research, the redox behavior and the charge transfer µΑ kinetics of DL -Leucine, L-Proline and L-Tryptophan in 0 the presence and absence of copper and iron will be investigated. Current ( -20 Experimental A computerized electrochemistry system developed by -40 -800 -400 0 400 800 Advanced Analytics, Virginia, USA, (Model-2040) Potential vs. Ag/AgCl (mV) consisting of three electrodes micro-cell with a saturated Ag/AgCl reference, a Pt-wire auxiliary and a pretreated Fig.2 . Comparison of the cyclic voltammogram of Pt-button working electrode is employed to investigate 5.0mM (a) L-Proline, (b) Cu-L-Proline and (c) Fe-L- Proline in 0.1M KCl solution at a Pt-button different amino acids and metal-amino acid systems. -

Effect of Ph on the Binding of Sodium, Lysine, and Arginine Counterions to L- Undecyl Leucinate Micelles
Effect of pH on the Binding of Sodium, Lysine, and Arginine Counterions to l- Undecyl Leucinate Micelles Corbin Lewis, Burgoyne H. Hughes, Mariela Vasquez, Alyssa M. Wall, Victoria L. Northrup, Tyler J. Witzleb, Eugene J. Billiot, et al. Journal of Surfactants and Detergents ISSN 1097-3958 Volume 19 Number 6 J Surfact Deterg (2016) 19:1175-1188 DOI 10.1007/s11743-016-1875-y 1 23 Your article is protected by copyright and all rights are held exclusively by AOCS. This e- offprint is for personal use only and shall not be self-archived in electronic repositories. If you wish to self-archive your article, please use the accepted manuscript version for posting on your own website. You may further deposit the accepted manuscript version in any repository, provided it is only made publicly available 12 months after official publication or later and provided acknowledgement is given to the original source of publication and a link is inserted to the published article on Springer's website. The link must be accompanied by the following text: "The final publication is available at link.springer.com”. 1 23 Author's personal copy J Surfact Deterg (2016) 19:1175–1188 DOI 10.1007/s11743-016-1875-y ORIGINAL ARTICLE Effect of pH on the Binding of Sodium, Lysine, and Arginine Counterions to L-Undecyl Leucinate Micelles 1 2 1 2 Corbin Lewis • Burgoyne H. Hughes • Mariela Vasquez • Alyssa M. Wall • 2 2 1 3 Victoria L. Northrup • Tyler J. Witzleb • Eugene J. Billiot • Yayin Fang • 1 2 Fereshteh H. Billiot • Kevin F. Morris Received: 20 May 2016 / Accepted: 6 September 2016 / Published online: 20 September 2016 Ó AOCS 2016 Abstract Micelle formation by the amino acid-based sur- surface through both of its amine functional groups. -

Amino Acid Chemistry
Handout 4 Amino Acid and Protein Chemistry ANSC 619 PHYSIOLOGICAL CHEMISTRY OF LIVESTOCK SPECIES Amino Acid Chemistry I. Chemistry of amino acids A. General amino acid structure + HN3- 1. All amino acids are carboxylic acids, i.e., they have a –COOH group at the #1 carbon. 2. All amino acids contain an amino group at the #2 carbon (may amino acids have a second amino group). 3. All amino acids are zwitterions – they contain both positive and negative charges at physiological pH. II. Essential and nonessential amino acids A. Nonessential amino acids: can make the carbon skeleton 1. From glycolysis. 2. From the TCA cycle. B. Nonessential if it can be made from an essential amino acid. 1. Amino acid "sparing". 2. May still be essential under some conditions. C. Essential amino acids 1. Branched chain amino acids (isoleucine, leucine and valine) 2. Lysine 3. Methionine 4. Phenyalanine 5. Threonine 6. Tryptophan 1 Handout 4 Amino Acid and Protein Chemistry D. Essential during rapid growth or for optimal health 1. Arginine 2. Histidine E. Nonessential amino acids 1. Alanine (from pyruvate) 2. Aspartate, asparagine (from oxaloacetate) 3. Cysteine (from serine and methionine) 4. Glutamate, glutamine (from α-ketoglutarate) 5. Glycine (from serine) 6. Proline (from glutamate) 7. Serine (from 3-phosphoglycerate) 8. Tyrosine (from phenylalanine) E. Nonessential and not required for protein synthesis 1. Hydroxyproline (made postranslationally from proline) 2. Hydroxylysine (made postranslationally from lysine) III. Acidic, basic, polar, and hydrophobic amino acids A. Acidic amino acids: amino acids that can donate a hydrogen ion (proton) and thereby decrease pH in an aqueous solution 1. -

COVID-19: the Disease, the Immunological Challenges, the Treatment with Pharmaceuticals and Low-Dose Ionizing Radiation
cells Review COVID-19: The Disease, the Immunological Challenges, the Treatment with Pharmaceuticals and Low-Dose Ionizing Radiation Jihang Yu 1 , Edouard I. Azzam 1, Ashok B. Jadhav 1 and Yi Wang 1,2,* 1 Radiobiology and Health, Isotopes, Radiobiology & Environment Directorate (IRED), Canadian Nuclear Laboratories (CNL), Chalk River, ON K0J 1J0, Canada; [email protected] (J.Y.); [email protected] (E.I.A.); [email protected] (A.B.J.) 2 Department of Biochemistry Microbiology and Immunology, Faculty of Medicine, University of Ottawa, Ottawa, ON K1H 8M5, Canada * Correspondence: [email protected]; Tel.: +1-613-584-3311 (ext. 42653) Abstract: The year 2020 will be carved in the history books—with the proliferation of COVID-19 over the globe and with frontline health workers and basic scientists worldwide diligently fighting to alleviate life-threatening symptoms and curb the spread of the disease. Behind the shocking prevalence of death are countless families who lost loved ones. To these families and to humanity as a whole, the tallies are not irrelevant digits, but a motivation to develop effective strategies to save lives. However, at the onset of the pandemic, not many therapeutic choices were available besides supportive oxygen, anti-inflammatory dexamethasone, and antiviral remdesivir. Low-dose radiation (LDR), at a much lower dosage than applied in cancer treatment, re-emerged after a Citation: Yu, J.; Azzam, E.I.; Jadhav, 75-year silence in its use in unresolved pneumonia, as a scientific interest with surprising effects in A.B.; Wang, Y. COVID-19: The soothing the cytokine storm and other symptoms in severe COVID-19 patients. -

Amino Acids Amino Acids
Amino Acids Amino Acids What Are Amino Acids? Essential Amino Acids Non Essential Amino Acids Amino acids are the building blocks of proteins; proteins are made of amino acids. Isoleucine Arginine (conditional) When you ingest a protein your body breaks it down into the individual aminos, Leucine Glutamine (conditional) reorders them, re-folds them, and turns them into whatever is needed by the body at Lysine Tyrosine (conditional) that time. From only 20 amino acids, the body is able to make thousands of unique proteins with different functions. Methionine Cysteine (conditional) Phenylalanine Glycine (conditional) Threonine Proline (conditional) Did You Know? Tryptophan Serine (conditional) Valine Ornithine (conditional) There are 20 different types of amino acids that can be combined to make a protein. Each protein consists of 50 to 2,000 amino acids that are connected together in a specific Histidine* Alanine sequence. The sequence of the amino acids determines each protein’s unique structure Asparagine and its specific function in the body. Asparate Popular Amino Acid Supplements How Do They Benefit Our Health? Acetyl L- Carnitine: As part of its role in supporting L-Lysine: L-Lysine, an essential amino acid, is mental function, Acetyl L-Carnitine may help needed to support proper growth and bone Proteins (amino acids) are needed by your body to maintain muscles, bones, blood, as support memory, attention span and mental development. It can also support immune function. well as create enzymes, neurotransmitters and antibodies, as well as transport and performance. store molecules. N-Acetyl Cysteine: N-Acetyl Cysteine (NAC) is a L-Arginine: L-Arginine is a nonessential amino acid form of the amino acid cysteine.