Amino Acid Chemistry
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Characterization of L-Serine Deaminases, Sdaa (PA2448) and Sdab 2 (PA5379), and Their Potential Role in Pseudomonas Aeruginosa 3 Pathogenesis 4 5 Sixto M
bioRxiv preprint doi: https://doi.org/10.1101/394957; this version posted August 20, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Characterization of L-serine deaminases, SdaA (PA2448) and SdaB 2 (PA5379), and their potential role in Pseudomonas aeruginosa 3 pathogenesis 4 5 Sixto M. Leal1,6, Elaine Newman2 and Kalai Mathee1,3,4,5 * 6 7 Author affiliations: 8 9 1Department of Biological Sciences, College of Arts Sciences and 10 Education, Florida International University, Miami, United States of 11 America 12 2Department of Biological Sciences, Concordia University, Montreal, 13 Canada 14 3Department of Molecular Microbiology and Infectious Diseases, Herbert 15 Wertheim College of Medicine, Florida International University, Miami, 16 United States of America 17 4Biomolecular Sciences Institute, Florida International University, Miami, 18 United States of America 19 20 Present address: 21 22 5Department of Human and Molecular Genetics, Herbert Wertheim 23 College of Medicine, Florida International University, Miami, United States 24 of America 25 6Case Western Reserve University, United States of America 26 27 28 *Correspondance: Kalai Mathee, MS, PhD, 29 [email protected] 30 31 Telephone : 1-305-348-0628 32 33 Keywords: Serine Catabolism, Central Metabolism, TCA Cycle, Pyruvate, 34 Leucine Responsive Regulatory Protein (LRP), One Carbon Metabolism 35 Running title: P. aeruginosa L-serine deaminases 36 Subject category: Pathogenicity and Virulence/Host Response 37 1 bioRxiv preprint doi: https://doi.org/10.1101/394957; this version posted August 20, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. -

Insights Into the Molecular Basis for Substrate Binding and Specificity of the Wild-Type L-Arginine/Agmatine Antiporter Adic
Insights into the molecular basis for substrate binding and specificity of the wild-type L-arginine/agmatine antiporter AdiC Hüseyin Ilgüa,b,1, Jean-Marc Jeckelmanna,b,1, Vytautas Gapsysc, Zöhre Ucuruma,b, Bert L. de Grootc, and Dimitrios Fotiadisa,b,2 aInstitute of Biochemistry and Molecular Medicine, University of Bern, CH-3012 Bern, Switzerland; bSwiss National Centre of Competence in Research TransCure, University of Bern, CH-3012 Bern, Switzerland; and cComputational Biomolecular Dynamics Group, Max-Planck-Institute for Biophysical Chemistry, D-37077 Goettingen, Germany Edited by Christopher Miller, Howard Hughes Medical Institute, Brandeis University, Waltham, MA, and approved July 26, 2016 (received for review April 4, 2016) Pathogenic enterobacteria need to survive the extreme acidity of Arg-bound states (10). The two outward-open, substrate-free the stomach to successfully colonize the human gut. Enteric bacteria structures are at the reasonable and moderate resolutions of 3.2 Å circumvent the gastric acid barrier by activating extreme acid- (8) and 3.6 Å (9), respectively, and the only ones available of wild- resistance responses, such as the arginine-dependent acid resistance type AdiC (AdiC-wt). The two other structures are with bound system. In this response, L-arginine is decarboxylated to agmatine, Arg and at 3-Å resolution, and could only be obtained as a result thereby consuming one proton from the cytoplasm. In Escherichia of the introduction of specific point mutations: AdiC-N22A (10) coli,theL-arginine/agmatine antiporter AdiC facilitates the export and AdiC-N101A (11). The N101A mutation results in a defective of agmatine in exchange of L-arginine, thus providing substrates for AdiC protein unable to bind Arg and with a dramatically de- further removal of protons from the cytoplasm and balancing the creased turnover rate compared with wild-type (11). -

Separation of Isomers <Emphasis Type="Italic">L </Emphasis>-Alanine and Sarcosine in Urine by Electrospra
SHORT COMMUNICATION Separation of Isomers L-Alanine and Sarcosine in Urine by Electrospray Ionization and Tandem Differential Mobility Analysis-Mass Spectrometry Pablo Martínez-Lozanoa and Juan Rusb a Institute for Biomedical Technologies-National Research Council, Milan, Italy b SEADM, Valladolid, Spain Sarcosine, an isomer of L-alanine, has been proposed as a prostate cancer progression biomarker [1]. Both compounds are detected in urine, where the measured sarcosine/alanine ratio has been found to be higher in prostate biopsy-positive group versus controls. We present here preliminary evidence showing that urine samples spiked with sarcosine/alanine can be partially resolved in 3 min via tandem differential mobility analysis-mass spectrometry (DMA-MS). Based on the calibration curves obtained for two mobility peaks, we finally estimate their concentration ratio in urine. (J Am Soc Mass Spectrom 2010, 21, 1129–1132) © 2010 American Society for Mass Spectrometry rostate cancer is a leading cause of death among the tive-ion mode at the DMA entrance slit. Our nanospray male population. Sarcosine, an isomer of alanine, parameters were: capillary 360 m o.d., 50 m i.d., length Pwas recently proposed as a prostate cancer progres- 22 cm, driving pressure 75 mbar, and 3.8 kV. The ions sion biomarker [1]. In particular, the sarcosine/alanine entered the separation region propelled by an electric field ratio was found to be significantly higher in urine derived and against a counterflow gas (0.2 L/min). Sheath gas from biopsy-positive prostate cancer patients compared flow was kept constant, and the classification voltage was with biopsy-negative controls. -

Effect of Peptide Histidine Isoleucine on Water and Electrolyte Transport in the Human Jejunum
Gut: first published as 10.1136/gut.25.6.624 on 1 June 1984. Downloaded from Gut, 1984, 25, 624-628 Alimentary tract and pancreas Effect of peptide histidine isoleucine on water and electrolyte transport in the human jejunum K J MORIARTY, J E HEGARTY, K TATEMOTO, V MUTT, N D CHRISTOFIDES, S R BLOOM, AND J R WOOD From the Department of Gastroenterology, St Bartholomew's Hospital, London, The Liver Unit, King's College Hospital, London, Department ofMedicine, Hammersmith Hospital, London, and Department of Biochemistry, Karolinska Institute, Stockholm, Sweden SUMMARY Peptide histidine isoleucine, a 27 amino acid peptide with close amino acid sequence homology to vasoactive intestinal peptide and secretin, is distributed throughout the mammalian intestinal tract, where it has been localised to intramural neurones. An intestinal perfusion technique has been used to study the effect of intravenous peptide histidine isoleucine (44.5 pmol/kg/min) on water and electrolyte transport from a plasma like electrolyte solution in human jejunum in vivo. Peptide histidine isoleucine infusion produced peak plasma peptide histidine isoleucine concentrations in the range 2000-3000 pmolIl, flushing, tachycardia and a reduction in diastolic blood pressure. Peptide histidine isoleucine caused a significant inhibition of net absorption of water, sodium, potassium and bicarbonate and induced a net secretion of chloride, these changes being completely reversed during the post-peptide histidine isoleucine period. These findings suggest that endogenous peptide histidine isoleucine may participate in the neurohumoral regulation of water and electrolyte transport in the human jejunum. http://gut.bmj.com/ Peptide histidine isoleucine, isolated originally from INTESTINAL PERFUSION mammalian small intestine, is a 27-amino acid After an eight hour fast, each subject swallowed a peptide having close amino acid sequence homology double lumen intestinal perfusion tube, incorpo- to vasoactive intestinal peptide and secretin. -

Isoleucine, an Essential Amino Acid, Prevents Liver Metastases of Colon Cancer by Antiangiogenesis Kazumoto Murata1 and Masami Moriyama2
Research Article Isoleucine, an Essential Amino Acid, Prevents Liver Metastases of Colon Cancer by Antiangiogenesis Kazumoto Murata1 and Masami Moriyama2 1The First Department of Internal Medicine, Mie University School of Medicine, Tsu, Mie, Japan and 2Microbiology and Immunology, Keio University School of Medicine, Tokyo, Japan Abstract infection in the airways of cystic fibrosis (8), and susceptibility to salmonella infection in mouse intestinal tracts (9). In addition to In spite of recent advances in the treatment of colon cancer, h multiple liver metastases of colon cancer are still difficult to their direct antimicrobial activities, -defensins are strong treat. Some chemotherapeutic regimens have been reported to chemotactic factors for memory T cells and dendritic cells, be efficient, but there is a high risk of side effects associated suggesting that they also play an important role in acquired immunity (10–12). h-defensins are also inducible by inflammatory with these. Here, we show that isoleucine, an essential amino a h acid, prevents liver metastases in a mouse colon cancer cytokines, such as tumor necrosis factor- and interleukin-1 (13, 14). Recently, Fehlbaum et al. (15) reported that isoleucine metastatic model. Because isoleucine is a strong inducer of h B-defensin, we first hypothesized that it prevented liver and its analogues are highly specific inducers of -defensins. We thus originally hypothesized that isoleucine may contribute to metastases via the accumulation of dendritic cells or memory B tumor immunity through both innate and acquired immunity by T cells through up-regulation of -defensin. However, neither h B-defensin nor immunologic responses were induced by induction of -defensins. -

The Role of Agmatine and Arginine Decarboxylase in Ischemic Tolerance After Transient Cerebral Ischemia in Rat Models
The role of agmatine and arginine decarboxylase in ischemic tolerance after transient cerebral ischemia in rat models Jin Young Jung Department of Medicine The Graduate School, Yonsei University The role of agmatine and arginine decarboxylase in ischemic tolerance after transient cerebral ischemia in rat models Directed by Professor Seung Kon Huh The Doctoral Dissertation submitted to the Department of Medicine, the Graduate School of Yonsei University in partial fulfillment of the requirements for the degree of Doctor of Philosophy Jin Young Jung May 2007 This certifies that the Doctoral Dissertation of Jin Young Jung is approved. __________________________________ Thesis Supervisor: Seung Kon Huh __________________________________ Jong Eun Lee: Thesis Committee Member #1 __________________________________ Jin Woo Chang: Thesis Committee Member #2 __________________________________ Duck Sun Ahn: Thesis Committee Member #3 __________________________________ Ji Cheol Shin: Thesis Committee Member #4 The Graduate School Yonsei University May 2007 Acknowledgements Some may consider this short section of the thesis trivial but for me it is a chance to express my sincerest gratitude to those that I am truly thankful. First of all, I would like to express my deepest gratitude to my thesis supervisor and mentor Professor Seung Kon Huh. He has inspired me when I was troubled and always gave me a warm heart. I would also like to thank Professor Jong Eun Lee who shared her valuable time on the execution and interpretation of this study, Professor Jin Woo Chang who always inspiring me with passion and discerning insight. Professor Duck Sun Ahn whose insightful comments were essential in completing this thesis, Professor Ji Cheol Shin for the excellent suggestion for improvement in this thesis. -
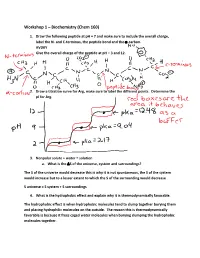
Workshop 1 – Biochemistry (Chem 160)
Workshop 1 – Biochemistry (Chem 160) 1. Draw the following peptide at pH = 7 and make sure to include the overall charge, label the N- and C-terminus, the peptide bond and the -carbon. AVDKY Give the overall charge of the peptide at pH = 3 and 12. 2. Draw a titration curve for Arg, make sure to label the different points. Determine the pI for Arg. 3. Nonpolar solute + water = solution a. What is the S of the universe, system and surroundings? The S of the universe would decrease this is why it is not spontaneous, the S of the system would increase but to a lesser extent to which the S of the surrounding would decrease S universe = S system + S surroundings 4. What is the hydrophobic effect and explain why it is thermodynamically favorable. The hydrophobic effect is when hydrophobic molecules tend to clump together burying them and placing hydrophilic molecules on the outside. The reason this is thermodynamically favorable is because it frees caged water molecules when burying clumping the hydrophobic molecules together. 5. Urea dissolves very readily in water, but the solution becomes very cold as the urea dissolves. How is this possible? Urea dissolves in water because when dissolving there is a net increase in entropy of the universe. The heat exchange, getting colder only reflects the enthalpy (H) component of the total energy change. The entropy change is high enough to offset the enthalpy component and to add up to an overall -G 6. A mutation that changes an alanine residue in the interior of a protein to valine is found to lead to a loss of activity. -

Effects of Single Amino Acid Deficiency on Mrna Translation Are Markedly
www.nature.com/scientificreports OPEN Efects of single amino acid defciency on mRNA translation are markedly diferent for methionine Received: 12 December 2016 Accepted: 4 May 2018 versus leucine Published: xx xx xxxx Kevin M. Mazor, Leiming Dong, Yuanhui Mao, Robert V. Swanda, Shu-Bing Qian & Martha H. Stipanuk Although amino acids are known regulators of translation, the unique contributions of specifc amino acids are not well understood. We compared efects of culturing HEK293T cells in medium lacking either leucine, methionine, histidine, or arginine on eIF2 and 4EBP1 phosphorylation and measures of mRNA translation. Methionine starvation caused the most drastic decrease in translation as assessed by polysome formation, ribosome profling, and a measure of protein synthesis (puromycin-labeled polypeptides) but had no signifcant efect on eIF2 phosphorylation, 4EBP1 hyperphosphorylation or 4EBP1 binding to eIF4E. Leucine starvation suppressed polysome formation and was the only tested condition that caused a signifcant decrease in 4EBP1 phosphorylation or increase in 4EBP1 binding to eIF4E, but efects of leucine starvation were not replicated by overexpressing nonphosphorylatable 4EBP1. This suggests the binding of 4EBP1 to eIF4E may not by itself explain the suppression of mRNA translation under conditions of leucine starvation. Ribosome profling suggested that leucine deprivation may primarily inhibit ribosome loading, whereas methionine deprivation may primarily impair start site recognition. These data underscore our lack of a full -

Production of L-Asparaginase II by Escherichia Coli HOWARD CEDAR and JAMES H
JOURNAL OF BACTERIOLOGY, Dec. 1968, p. 2043-2048 Vol. 96, No. 6 Copyright @ 1968 American Society for Microbiology Printed in U.S.A. Production of L-Asparaginase II by Escherichia coli HOWARD CEDAR AND JAMES H. SCHWARTZ Department of Microbiology, New York University School ofMedicine, New York, New York 10016 Received for publication 30 July 1968 L-Asparaginase II was synthesized at constant rates by Escherichia coli under anaerobic conditions. The enzyme was produced optimally by bacteria grown between pH 7 and 8 at 37 C. Although some enzyme was formed aerobically, be- tween 100 and 1,000 times more asparaginase II was produced during anaerobic growth in media enriched with high concentrations of a variety of amino acids. Bacteria grown under these conditions should provide a rich starting material for the large-scale production of the enzyme. No single amino acid specifically induced the synthesis of the asparaginase, nor did L-asparagine, even when it was used as the only source of nitrogen. The enzyme was produced at lower rates in the presence of sugars; glucose was the most inhibitory. Deamidation of L-asparagine by extracts of the conditions which control the production of Escherichia coli was first reported in 1957 by asparaginase II. Tsuji (28). E. coli was later shown to produce two distinct asparaginases (L-asparagine amido- MATERIALS AND METHODS hydrolase, EC 3.5.1 .1) which differ in a number For most of the experiments, we used E. coli K-12 of properties, perhaps most significantly in wild type. We also used strain 22-64, which lacks their markedly different affinities for asparagine citrate synthase (10), and strain 309-1, which lacks (5, 24, 27). -

Beta Alanine
PERFORMANCE ENHANCERS FACTS AND BOTTOM LINE BETA ALANINE What is it? B-alanine is a naturally occurring amino acid (a non-essential amino acid) not used by the body to make muscle tissue. Rather, research has shown that B-alanine works by increasing the muscle content of an important compound – carnosine. In fact, the production of carnosine is limited by the availability of B-alanine. Carnosine is highly concentrated in muscle tissue where its role is primarily to soak up hydrogen ions. Does it work? B-alanine is one of the few dietary supplements that actually have good scientific evidence that it can possibly enhance performance. How does it work? When you exercise intensely the body produces hydrogen ions. The longer you exercise the more hydrogen ions you produce and this reduces the pH level in your muscles. Muscles work best in a very specific pH range and when the pH drops below that level then muscular performance also starts to decrease. Anything that helps to prevent or delay that drop in pH will help delay muscle fatigue. This is where B-alanine has proven to be very helpful. Beta-alanine increases the levels of carnosine in your slow and fast twitch muscle fibers and carnosine is a buffer that basically soaks up hydrogen ions and so reduces the drop in pH. By keeping your hydrogen ion levels lower, B-alanine allows you to train harder and longer. The bottom line is that B-alanine works by increasing the hydrogen ion buffering abilities of your muscles. What benefits does it offer? B-alanine has been shown to increase muscle strength, increase muscle mass, increase anaerobic endurance, increase aerobic endurance and increase exercise capacity. -

Amino Acid Recognition by Aminoacyl-Trna Synthetases
www.nature.com/scientificreports OPEN The structural basis of the genetic code: amino acid recognition by aminoacyl‑tRNA synthetases Florian Kaiser1,2,4*, Sarah Krautwurst3,4, Sebastian Salentin1, V. Joachim Haupt1,2, Christoph Leberecht3, Sebastian Bittrich3, Dirk Labudde3 & Michael Schroeder1 Storage and directed transfer of information is the key requirement for the development of life. Yet any information stored on our genes is useless without its correct interpretation. The genetic code defnes the rule set to decode this information. Aminoacyl-tRNA synthetases are at the heart of this process. We extensively characterize how these enzymes distinguish all natural amino acids based on the computational analysis of crystallographic structure data. The results of this meta-analysis show that the correct read-out of genetic information is a delicate interplay between the composition of the binding site, non-covalent interactions, error correction mechanisms, and steric efects. One of the most profound open questions in biology is how the genetic code was established. While proteins are encoded by nucleic acid blueprints, decoding this information in turn requires proteins. Te emergence of this self-referencing system poses a chicken-or-egg dilemma and its origin is still heavily debated 1,2. Aminoacyl-tRNA synthetases (aaRSs) implement the correct assignment of amino acids to their codons and are thus inherently connected to the emergence of genetic coding. Tese enzymes link tRNA molecules with their amino acid cargo and are consequently vital for protein biosynthesis. Beside the correct recognition of tRNA features3, highly specifc non-covalent interactions in the binding sites of aaRSs are required to correctly detect the designated amino acid4–7 and to prevent errors in biosynthesis5,8. -

Introduction to Proteins and Amino Acids Introduction
Introduction to Proteins and Amino Acids Introduction • Twenty percent of the human body is made up of proteins. Proteins are the large, complex molecules that are critical for normal functioning of cells. • They are essential for the structure, function, and regulation of the body’s tissues and organs. • Proteins are made up of smaller units called amino acids, which are building blocks of proteins. They are attached to one another by peptide bonds forming a long chain of proteins. Amino acid structure and its classification • An amino acid contains both a carboxylic group and an amino group. Amino acids that have an amino group bonded directly to the alpha-carbon are referred to as alpha amino acids. • Every alpha amino acid has a carbon atom, called an alpha carbon, Cα ; bonded to a carboxylic acid, –COOH group; an amino, –NH2 group; a hydrogen atom; and an R group that is unique for every amino acid. Classification of amino acids • There are 20 amino acids. Based on the nature of their ‘R’ group, they are classified based on their polarity as: Classification based on essentiality: Essential amino acids are the amino acids which you need through your diet because your body cannot make them. Whereas non essential amino acids are the amino acids which are not an essential part of your diet because they can be synthesized by your body. Essential Non essential Histidine Alanine Isoleucine Arginine Leucine Aspargine Methionine Aspartate Phenyl alanine Cystine Threonine Glutamic acid Tryptophan Glycine Valine Ornithine Proline Serine Tyrosine Peptide bonds • Amino acids are linked together by ‘amide groups’ called peptide bonds.