The Role of Reduced Methionine in Mediating the Metabolic Responses to Protein Restriction Using Different Sources of Protein
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
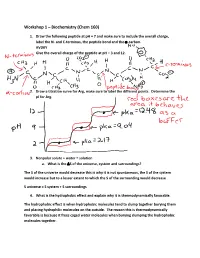
Workshop 1 – Biochemistry (Chem 160)
Workshop 1 – Biochemistry (Chem 160) 1. Draw the following peptide at pH = 7 and make sure to include the overall charge, label the N- and C-terminus, the peptide bond and the -carbon. AVDKY Give the overall charge of the peptide at pH = 3 and 12. 2. Draw a titration curve for Arg, make sure to label the different points. Determine the pI for Arg. 3. Nonpolar solute + water = solution a. What is the S of the universe, system and surroundings? The S of the universe would decrease this is why it is not spontaneous, the S of the system would increase but to a lesser extent to which the S of the surrounding would decrease S universe = S system + S surroundings 4. What is the hydrophobic effect and explain why it is thermodynamically favorable. The hydrophobic effect is when hydrophobic molecules tend to clump together burying them and placing hydrophilic molecules on the outside. The reason this is thermodynamically favorable is because it frees caged water molecules when burying clumping the hydrophobic molecules together. 5. Urea dissolves very readily in water, but the solution becomes very cold as the urea dissolves. How is this possible? Urea dissolves in water because when dissolving there is a net increase in entropy of the universe. The heat exchange, getting colder only reflects the enthalpy (H) component of the total energy change. The entropy change is high enough to offset the enthalpy component and to add up to an overall -G 6. A mutation that changes an alanine residue in the interior of a protein to valine is found to lead to a loss of activity. -

Effects of Basic Amino Acids and Their Derivatives on SARS-Cov-2 and Influenza-A Virus Infection
viruses Article Effects of Basic Amino Acids and Their Derivatives on SARS-CoV-2 and Influenza-A Virus Infection Ivonne Melano 1 , Li-Lan Kuo 2, Yan-Chung Lo 3, Po-Wei Sung 4, Ni Tien 5,6 and Wen-Chi Su 1,2,7,* 1 Graduate Institute of Biomedical Sciences, College of Medicine, China Medical University, Taichung 40402, Taiwan; [email protected] 2 Research Center for Emerging Viruses, China Medical University Hospital, Taichung 40402, Taiwan; [email protected] 3 Sinphar Pharmaceutical Co., Ltd., Sinphar Group, Yilan 269, Taiwan; [email protected] 4 School of Medicine, China Medical University, Taichung 40402, Taiwan; [email protected] 5 Department of Laboratory Medicine, China Medical University Hospital, Taichung 40402, Taiwan; [email protected] 6 Department of Medical Laboratory Science and Biotechnology, China Medical University, Taichung 40402, Taiwan 7 International Master’s Program of Biomedical Sciences, China Medical University, Taichung 40402, Taiwan * Correspondence: [email protected] Abstract: Amino acids have been implicated with virus infection and replication. Here, we demon- strate the effects of two basic amino acids, arginine and lysine, and their ester derivatives on infection of two enveloped viruses, SARS-CoV-2, and influenza A virus. We found that lysine and its ester derivative can efficiently block infection of both viruses in vitro. Furthermore, the arginine ester derivative caused a significant boost in virus infection. Studies on their mechanism of action revealed that the compounds potentially disturb virus uncoating rather than virus attachment and endosomal Citation: Melano, I.; Kuo, L.-L.; Lo, acidification. Our findings suggest that lysine supplementation and the reduction of arginine-rich Y.-C.; Sung, P.-W.; Tien, N.; Su, W.-C. -

Amino Acid Chemistry
Handout 4 Amino Acid and Protein Chemistry ANSC 619 PHYSIOLOGICAL CHEMISTRY OF LIVESTOCK SPECIES Amino Acid Chemistry I. Chemistry of amino acids A. General amino acid structure + HN3- 1. All amino acids are carboxylic acids, i.e., they have a –COOH group at the #1 carbon. 2. All amino acids contain an amino group at the #2 carbon (may amino acids have a second amino group). 3. All amino acids are zwitterions – they contain both positive and negative charges at physiological pH. II. Essential and nonessential amino acids A. Nonessential amino acids: can make the carbon skeleton 1. From glycolysis. 2. From the TCA cycle. B. Nonessential if it can be made from an essential amino acid. 1. Amino acid "sparing". 2. May still be essential under some conditions. C. Essential amino acids 1. Branched chain amino acids (isoleucine, leucine and valine) 2. Lysine 3. Methionine 4. Phenyalanine 5. Threonine 6. Tryptophan 1 Handout 4 Amino Acid and Protein Chemistry D. Essential during rapid growth or for optimal health 1. Arginine 2. Histidine E. Nonessential amino acids 1. Alanine (from pyruvate) 2. Aspartate, asparagine (from oxaloacetate) 3. Cysteine (from serine and methionine) 4. Glutamate, glutamine (from α-ketoglutarate) 5. Glycine (from serine) 6. Proline (from glutamate) 7. Serine (from 3-phosphoglycerate) 8. Tyrosine (from phenylalanine) E. Nonessential and not required for protein synthesis 1. Hydroxyproline (made postranslationally from proline) 2. Hydroxylysine (made postranslationally from lysine) III. Acidic, basic, polar, and hydrophobic amino acids A. Acidic amino acids: amino acids that can donate a hydrogen ion (proton) and thereby decrease pH in an aqueous solution 1. -

Amino Acids Amino Acids
Amino Acids Amino Acids What Are Amino Acids? Essential Amino Acids Non Essential Amino Acids Amino acids are the building blocks of proteins; proteins are made of amino acids. Isoleucine Arginine (conditional) When you ingest a protein your body breaks it down into the individual aminos, Leucine Glutamine (conditional) reorders them, re-folds them, and turns them into whatever is needed by the body at Lysine Tyrosine (conditional) that time. From only 20 amino acids, the body is able to make thousands of unique proteins with different functions. Methionine Cysteine (conditional) Phenylalanine Glycine (conditional) Threonine Proline (conditional) Did You Know? Tryptophan Serine (conditional) Valine Ornithine (conditional) There are 20 different types of amino acids that can be combined to make a protein. Each protein consists of 50 to 2,000 amino acids that are connected together in a specific Histidine* Alanine sequence. The sequence of the amino acids determines each protein’s unique structure Asparagine and its specific function in the body. Asparate Popular Amino Acid Supplements How Do They Benefit Our Health? Acetyl L- Carnitine: As part of its role in supporting L-Lysine: L-Lysine, an essential amino acid, is mental function, Acetyl L-Carnitine may help needed to support proper growth and bone Proteins (amino acids) are needed by your body to maintain muscles, bones, blood, as support memory, attention span and mental development. It can also support immune function. well as create enzymes, neurotransmitters and antibodies, as well as transport and performance. store molecules. N-Acetyl Cysteine: N-Acetyl Cysteine (NAC) is a L-Arginine: L-Arginine is a nonessential amino acid form of the amino acid cysteine. -

Neurotransmitters: Critical Amino Acids Affecting
1 NEUROTRANSMITTERS: CRITICAL AMINO ACIDS AFFECTING SEROTONIN AND DOPAMINE Approved _____________________________Date: _August 19, 2015__________________ 2 NEUROTRANSMITTERS: CRITICAL AMINO ACIDS AFFECTING SEROTONIN AND DOPAMINE __________________ Seminar Paper Presented to The Graduate Faculty University of Wisconsin-Platteville __________________ In Partial Fulfillment of the Requirement for the Degree Masters of Science In Adult Education With Emphasis in Counseling __________________ By Laurie Williams 2015 3 Acknowledgments I am grateful to have had the privilege to work with many exceptional professors in their fields of study. I am particularly appreciative that I received both classroom training and advising from my program coordinator and advisor to my Seminar Paper, Dr. Patricia Bromley. I am very thankful for the efforts of all the professors and staff I worked with at UW-Platteville, as well as UW-Whitewater and UW-Rock County for my superior undergraduate and master’s education. I appreciate the opportunity and support to individually examine scholarly research including published articles, academic books, and professional books on my subjects of interest. 4 iii Abstract NEUROTRANSMITTERS: CRITICAL AMINO ACIDS AFFECTING SEROTONIN AND DOPAMINE This paper examines how amino acids from foods critically affect neurotransmitters, which, in turn, affect every area of brain and body health. The manner in which neurotransmitters can affect personality, mental health, and learning disabilities is explored in this research review. Inadequate amino acid nutrition or exposure to poisonous chemicals or drugs can directly cause nerve cells to function poorly. Resulting changes in quantities of neurotransmitters such as serotonin and dopamine can influence emotional states like anger, frustration, or depression, but can also influence energy, speed of thought, activity level, speech volume and fluency, social functioning, and memory. -

Nucleotide Base Coding and Am1ino Acid Replacemients in Proteins* by Emil L
VOL. 48, 1962 BIOCHEMISTRY: E. L. SAIITH 677 18 Britten, R. J., and R. B. Roberts, Science, 131, 32 (1960). '9 Crestfield, A. M., K. C. Smith, and F. WV. Allen, J. Biol. Chem., 216, 185 (1955). 20 Gamow, G., Nature, 173, 318 (1954). 21 Brenner, S., these PROCEEDINGS, 43, 687 (1957). 22 Nirenberg, M. WV., J. H. Matthaei, and 0. WV. Jones, unpublished data. 23 Crick, F. H. C., L. Barnett, S. Brenner, and R. J. Watts-Tobin, Nature, 192, 1227 (1961). 24 Levene, P. A., and R. S. Tipson, J. Biol. Ch-nn., 111, 313 (1935). 25 Gierer, A., and K. W. Mundry, Nature, 182, 1437 (1958). 2' Tsugita, A., and H. Fraenkel-Conrat, J. Mllot. Biol., in press. 27 Tsugita, A., and H. Fraenkel-Conrat, personal communication. 28 Wittmann, H. G., Naturwissenschaften, 48, 729 (1961). 29 Freese, E., in Structure and Function of Genetic Elements, Brookhaven Symposia in Biology, no. 12 (1959), p. 63. NUCLEOTIDE BASE CODING AND AM1INO ACID REPLACEMIENTS IN PROTEINS* BY EMIL L. SMITHt LABORATORY FOR STUDY OF HEREDITARY AND METABOLIC DISORDERS AND THE DEPARTMENTS OF BIOLOGICAL CHEMISTRY AND MEDICINE, UNIVERSITY OF UTAH COLLEGE OF MEDICINE Communicated by Severo Ochoa, February 14, 1962 The problem of which bases of messenger or template RNA' specify the coding of amino acids in proteins has been largely elucidated by the use of synthetic polyri- bonucleotides.2-7 For these triplet nucleotide compositions (Table 1), it is of in- terest to examine some of the presently known cases of amino acid substitutions in polypeptides or proteins of known structure. -

Recent Progress in the Microbial Production of Pyruvic Acid
fermentation Review Recent Progress in the Microbial Production of Pyruvic Acid Neda Maleki 1 and Mark A. Eiteman 2,* 1 Department of Food Science, Engineering and Technology, University of Tehran, Karaj 31587-77871, Iran; [email protected] 2 School of Chemical, Materials and Biomedical Engineering, University of Georgia, Athens, GA 30602, USA * Correspondence: [email protected]; Tel.: +1-706-542-0833 Academic Editor: Gunnar Lidén Received: 10 January 2017; Accepted: 6 February 2017; Published: 13 February 2017 Abstract: Pyruvic acid (pyruvate) is a cellular metabolite found at the biochemical junction of glycolysis and the tricarboxylic acid cycle. Pyruvate is used in food, cosmetics, pharmaceutical and agricultural applications. Microbial production of pyruvate from either yeast or bacteria relies on restricting the natural catabolism of pyruvate, while also limiting the accumulation of the numerous potential by-products. In this review we describe research to improve pyruvate formation which has targeted both strain development and process development. Strain development requires an understanding of carbohydrate metabolism and the many competing enzymes which use pyruvate as a substrate, and it often combines classical mutation/isolation approaches with modern metabolic engineering strategies. Process development requires an understanding of operational modes and their differing effects on microbial growth and product formation. Keywords: auxotrophy; Candida glabrata; Escherichia coli; fed-batch; metabolic engineering; pyruvate; pyruvate dehydrogenase 1. Introduction Pyruvic acid (pyruvate at neutral pH) is a three carbon oxo-monocarboxylic acid, also known as 2-oxopropanoic acid, 2-ketopropionic acid or acetylformic acid. Pyruvate is biochemically located at the end of glycolysis and entry into the tricarboxylic acid (TCA) cycle (Figure1). -

Amino Acid Transport Pathways in the Small Intestine of the Neonatal Rat
Pediat. Res. 6: 713-719 (1972) Amino acid neonate intestine transport, amino acid Amino Acid Transport Pathways in the Small Intestine of the Neonatal Rat J. F. FITZGERALD1431, S. REISER, AND P. A. CHRISTIANSEN Departments of Pediatrics, Medicine, and Biochemistry, and Gastrointestinal Research Laboratory, Indiana University School of Medicine and Veterans Administration Hospital, Indianapolis, Indiana, USA Extract The activity of amino acid transport pathways in the small intestine of the 2-day-old rat was investigated. Transport was determined by measuring the uptake of 1 mM con- centrations of various amino acids by intestinal segments after a 5- or 10-min incuba- tion and it was expressed as intracellular accumulation. The neutral amino acid transport pathway was well developed with intracellular accumulation values for leucine, isoleucine, valine, methionine, tryptophan, phenyl- alanine, tyrosine, and alanine ranging from 3.9-5.6 mM/5 min. The intracellular accumulation of the hydroxy-containing neutral amino acids threonine (essential) and serine (nonessential) were 2.7 mM/5 min, a value significantly lower than those of the other neutral amino acids. The accumulation of histidine was also well below the level for the other neutral amino acids (1.9 mM/5 min). The basic amino acid transport pathway was also operational with accumulation values for lysine, arginine and ornithine ranging from 1.7-2.0 mM/5 min. Accumulation of the essential amino acid lysine was not statistically different from that of nonessential ornithine. Ac- cumulation of aspartic and glutamic acid was only 0.24-0.28 mM/5 min indicating a very low activity of the acidic amino acid transport pathway. -

Meat Proteins in Human Nutrition
MEAT P80T€IN'S IN HUMAN NUTRITION J. KASTELIC Before I can begin to discuss the many questions which are implied in the title of this discussion I must beg your forbearance. I must clearly specify that I am appearing before you as a student of protein nutrition and not as an authority. This is not meant to be an apology. It is a statement which is not only pmdeut but entire1.y appropriate. And I must clear a bit of underb-msli amy before I give the su'oject before us specific consideration, ;^or I can be certain none of you came here only to listen to me reiterate the resalts of nitrogen balance studies published in the literature or to look at relmoductions of tables of data and of graphs which describe the results of the many biological value determinations of proteins that have been obt,ained frola laboratory animal experimentation with which so many of you are now familiar. It would be equally superfluous to call attention to the tremendous volume of literature that is now being published about the underlying eco- nomic, social, technological and medical problems which are confronting man in so many parts of the world today where food production is only sufficient to mainkin life. !here we were once so provincially concerned with vitamin defi- ciency diseases, we now appear to be rather singularly concerned about pro- tein malnutrition and how m might best cope with protein malnutrition in the human obliged to subsist on diets composed of protein-deficient foods. If there is still some controversy about the relative merits of proteins from plant and animal sources in 'numan nutrition it must stem from vexing economic considerations, meat animal production capabilities and problems associated with processing and storage; not fmm a lack of an appreciation of the nutritional role and functions of the amino acids which are essential to life. -

Protein and Amino Acid Requirements for Poultry by Todd J
PURDUE EXTENSION Animal Sciences AS-584-W Protein and Amino Acid Requirements for Poultry by Todd J. Applegate - Purdue University Roselina Angel – University of Maryland, College Park Introduction Essential amino acids must be supplied by the diet, This fact sheet has been developed to support the and a sufficient amount of non-essential amino acids implementation of the Natural Resources Conservation must also be supplied to prevent the conversion of Service Feed Management 592 Practice Standard. essential amino acids into non-essential amino acid. The Feed Management 592 Practice Standard was Additionally, if the amino acids supplied are not in adopted by NRCS in 2003 as another tool to assist the proper, or ideal, ratio in relation to the needs of with addressing resource concerns on livestock and the animal, then amino acids in excess of the least poultry operations. Feed management can assist with limiting amino acid will be deaminated and likely used reducing the import of nutrients to the farm and reduce as a source of energy rather than toward body protein the excretion of nutrients in manure. synthesis. This breakdown of amino acids will also Dietary crude protein (CP) requirements are result in higher nitrogenous excretions. somewhat of a misnomer as the requirement is The best way to reduce N in poultry excreta is to based on the amino acids content of the protein. lower the amount of CP that is fed by supplementing Once digested and absorbed, amino acids are used diets with amino acids. Reductions in the non-essential as the building blocks of structural proteins (muscle, amino acid pool, coupled with supplying a more skin, ligaments), metabolic proteins, enzymes, and “ideal” amino acid profile in the diet can substantially precursors of several body components. -

Radhakrishnan, Iii 4
NATURE OF THE GENETIC BLOCKS IN THE ISOLEUCINE-VALINE MUTANTS OF SALMONELLA R. P. WAGNER AND ARLOA BERGQUIST The Genetics Laboratory of the Department of Zoology, The Uniuersity of Texas, Austin, Texas Received April 26, 1960 HE metabolic pathways leading to the biosynthesis of isoleucine and valine are now sufficiently well understood, as a result of the work of a number of investigators ( STRASSMAN,THOMAS and WEINHOUSE1955; STRASSMAN,THOMAS, LOCKEand WEINHOUSE1956; STRASSMAN,SHATTON, CORSEY and WEINHOUSE 1958; UMBARGER1958a,b; ADELBERG1955; WAGNER,RADHAKRISHNAN and SNELL1958; RADHAKRISHNAN,WAGNER and SNELL1960; and RADHAKRISHNAN and SNELL1960) to make it possible to investigate the nature of the genetic blocks in mutant organisms requiring isoleucine and valine. This communication describes the results of the investigation of a series of mutants of Salmonella typhimurium originally isolated in the laboratory of DR. M. DEMEREC,The Carnegie Institute of Washington, Cold Spring Harbor, New York. It is limited to the purely biochemical aspects of these mutants, but is ,preceded by a com- munication from GLANVILLEand DEMEREC1960, which describes the linkage studies made with these mutants, and correlates the genetic with the biochemical 3ata. The biosynthesis of isoleucine and valine is believed to occur as shown in Figure 1. In the work to be described here only the steps proceeding from the 3-keto acids, a-acetolactic acid and a-aceto-P-hydroxybutyricacid, have been :onsidered in detail. The following abbreviations are used in Figure I and in mbsequent parts of this communication: AHB = a-aceto-a-hydroxybutyric acid; CHa C Ha CHa CHa I 1 I I c*o CHa-C-OH CHfC-OH CHI-C-H 1 I TPNH 1 I c.0 ~ *Valine I COOH COOH I COOH I COOH (PYRUVIC ACID) (ALI I (HKVI I (DHV) I I I I I I I I + Pyruvic Acid Step1 StepII Stepm Step H 4 I I I I cn3 CH, I CH3 I I I I I CHZ C=O I CH~CH~C-OH Threonine-GO I ~CH3CH2-~-OH-C=0I+! I I I I COOH COOH COOH COOH COOH (U-kmtobutyric acid1 (AH01 (HKII (DUI) (KI) FIGURE1 .-The biosynthetic pathway leading to isoleucine and valine. -

Role and Characteristics of Selected Amino Acid and Peptide Transporters in Epithelial Cells
TECHNISCHE UNIVERSITÄT MÜNCHEN Lehrstuhl für Ernährungsphysiologie Role and characteristics of selected amino acid and peptide transporters in epithelial cells Alexander Georg Nickel Vollständiger Abdruck der von der Fakultät Wissenschaftszentrum Weihenstephan für Ernährung, Landnutzung und Umwelt der Technischen Universität München zur Erlangung des akademischen Grades eines Doktors der Naturwissenschaften genehmigten Dissertation. Vorsitzender: Univ.-Prof. Dr. M. Schemann Prüfer der Dissertation: 1. Univ.-Prof. Dr. H. Daniel 2. Univ.-Prof. Dr. Th. F. Hofmann Die Dissertation wurde am 12.05.2009 bei der Technischen Universität München eingereicht und durch die Fakultät Wissenschaftszentrum Weihenstephan für Ernährung, Landnutzung und Umwelt am 01.09.2009 angenommen. A 2 Zum Erfolg braucht der Forscher die vier großen "G": Geist, Geduld, Geld und Glück. Paul Ehrlich 3 Table of contents 1 Characteristics of L-proline transport in OK cells................................................................ 10 1.1 Introduction ............................................................................................................. 10 1.1.1 Physiological importance of amino acids..................................................... 10 1.1.2 Basic principles of amino acid transport...................................................... 10 1.1.3 Amino acid transport in kidney .................................................................... 11 1.1.3.1 Apical amino acid transporters of the kidney proximal tubule ..................... 12 1.1.3.2